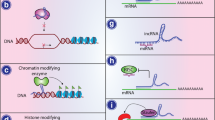
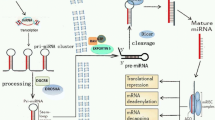
Abstract
Multiple sclerosis (MS) is a central nervous system chronic neuroinflammatory disease followed by neurodegeneration. The diagnosis is based on clinical presentation, cerebrospinal fluid testing and magnetic resonance imagining. There is still a lack of a diagnostic blood-based biomarker for MS. Due to the cost and difficulty of diagnosis, new and more easily accessible methods are being sought. New biomarkers should also allow for early diagnosis. Additionally, the treatment of MS should lead to the personalization of the therapy. MicroRNAs (miRNAs) and long non-coding RNAs (lncRNAs) as well as their target genes participate in pathophysiology processes in MS. Although the detailed mechanism of action of non-coding RNAs (ncRNAs, including miRNAs and lncRNAs) on neuroinflammation in MS has not been fully explained, several studies were conducted aiming to analyse their impact in MS. In this article, we review up-to-date knowledge on the latest research concerning the ncRNAs in MS and evaluate their role in neuroinflammation. We also point out the most promising ncRNAs which may be promising in MS as diagnostic and prognostic biomarkers.



Similar content being viewed by others
Availability of data and materials
The datasets generated and analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- ALS:
-
Amyotrophic lateral sclerosis
- APC:
-
Antigen-presenting cells
- BBB:
-
Blood-brain barrier
- BDMCs:
-
Bone marrow-derived dendritic cells
- BMDMs:
-
Bone marrow-derived macrophages
- CCL2:
-
C-C Motif Chemokine Ligand 2
- CCR6:
-
C-C Chemokine receptor 6
- CDMS:
-
Clinically definite MS
- CIS:
-
Clinically isolated syndrome
- CNS:
-
Central nervous system
- CSF:
-
Cerebrospinal fluid
- CXCL1:
-
C-X-C Motif Chemokine Ligand 1
- CXCL10:
-
C-X-C Motif Chemokine Ligand 10
- CXCL13:
-
C-X-C Motif chemokine 13
- dCLNs:
-
Deep cervical lymph nodes
- DA:
-
Dark Agouti
- DCs:
-
Dendritic cells
- EAE:
-
Experimental autoimmune encephalomyelitis
- EVs:
-
Extracellular vesicles
- FACS:
-
Fluorescence-activated Cell Sorting
- ECM:
-
Extracellular matrix
- SELE:
-
E-selectin
- GM-CSF:
-
Granulocyte Macrophage Colony- Stimulating Factor
- IgG:
-
Immunoglobulin G
- IL:
-
Interleukin
- KO:
-
Knockout
- LPS:
-
Lipopolysaccharide
- LincRNA:
-
Long intergenic non-coding RNA
- LncRNA:
-
Long non-coding RNA
- miR, miRNA:
-
MicroRNA
- MALAT1:
-
Metastasis associated lung adenocarcinoma transcript 1
- MMP9:
-
Matrix metalloproteinase 9
- MOG:
-
Myelin oligodendrocyte glycoprotein
- MRC1:
-
Mannose Receptor C-Type 1
- MRI:
-
Magnetic resonance imaging
- MS:
-
Multiple sclerosis
- MSCs:
-
Mesenchymal stem cells
- n:
-
Number
- ncRNAs:
-
non-coding RNAs
- OPN:
-
Osteopontin
- PBMC:
-
Peripheral blood mononuclear cells
- p.i.:
-
Post-immunization
- PPMS:
-
Primary progressive MS
- PVG:
-
Piebald Virol Glaxo
- RIS:
-
Radiologically isolated syndrome
- ROS:
-
Reactive oxygen species
- RRMS:
-
Relapsing-remitting MS
- Smad7:
-
Suppressor of mothers against decapentaplegic 7
- SOCS:
-
Suppressor of cytokine signalling proteins
- Th17:
-
T helper 17
- TNFα:
-
Tumour necrosis factor α
- WT:
-
Wild type
References
Thompson AJ, Banwell BL, Barkhof F et al (2018) Diagnosis of multiple sclerosis: 2017 revisions of the McDonald criteria. Lancet Neurol 17:162–173. https://doi.org/10.1016/S1474-4422(17)30470-2
Cordes KR, Srivastava D (2009) MicroRNA Regulation of Cardiovascular Development. Circ Res 104:724–732
Pordzik J, Jakubik D, Jarosz-Popek J et al (2019) Significance of circulating microRNAs in diabetes mellitus type 2 and platelet reactivity: bioinformatic analysis and review. Cardiovasc Diabetol 18:113. https://doi.org/10.1186/s12933-019-0918-x
Jakubik D, Fitas A, Eyileten C et al (2021) MicroRNAs and long non-coding RNAs in the pathophysiological processes of diabetic cardiomyopathy: emerging biomarkers and potential therapeutics. Cardiovasc Diabetol 20:55. https://doi.org/10.1186/s12933-021-01245-2
Zareba L, Fitas A, Wolska M et al (2020) MicroRNAs and Long Noncoding RNAs in Coronary Artery Disease: New and Potential Therapeutic Targets. Cardiol Clin 38:601–617. https://doi.org/10.1016/j.ccl.2020.07.005
Eyileten C, Wicik Z, De Rosa S et al (2018) MicroRNAs as Diagnostic and Prognostic Biomarkers in Ischemic Stroke-A Comprehensive Review and Bioinformatic Analysis. Cells 7. https://doi.org/10.3390/cells7120249
Sabatino J, Wicik Z, De Rosa S et al (2019) MicroRNAs fingerprint of bicuspid aortic valve. J Mol Cell Cardiol 134:98–106. https://doi.org/10.1016/j.yjmcc.2019.07.001
Pordzik J, Pisarz K, De Rosa S et al (2018) The potential role of platelet-related microRNAs in the development of cardiovascular events in high-risk populations, including diabetic patients: a review. Front Endocrinol 9:74. https://doi.org/10.3389/fendo.2018.00074
Wolska M, Jarosz-Popek J, Junger E et al (2020) Long Non-coding RNAs as Promising Therapeutic Approach in Ischemic Stroke: a Comprehensive Review. Mol Neurobiol. https://doi.org/10.1007/s12035-020-02206-8
Eyileten C, Sharif L, Wicik Z et al (2020) The Relation of the Brain-Derived Neurotrophic Factor with MicroRNAs in Neurodegenerative Diseases and Ischemic Stroke. Mol Neurobiol. https://doi.org/10.1007/s12035-020-02101-2
Jarosz-Popek J, Wolska M, Gasecka A et al (2020) The Importance of Non-Coding RNAs in Neurodegenerative Processes of Diabetes-Related Molecular Pathways. J Clin Med Res 10. https://doi.org/10.3390/jcm10010009
Pordzik J, Eyileten-Postuła C, Jakubik D et al (2021) MiR-126 Is an Independent Predictor of Long-Term All-Cause Mortality in Patients with Type 2 Diabetes Mellitus. J Clin Med Res 10. https://doi.org/10.3390/jcm10112371
Lukiw WJ, Handley P, Wong L, Crapper McLachlan DR (1992) BC200 RNA in normal human neocortex, non-Alzheimer dementia (NAD), and senile dementia of the Alzheimer type (AD). Neurochem Res 17:591–597. https://doi.org/10.1007/BF00968788
Ghoveud E, Teimuri S, Vatandoost J et al (2020) Potential Biomarker and Therapeutic LncRNAs in Multiple Sclerosis Through Targeting Memory B Cells. NeuroMol Med 22:111–120
Regev K, Healy BC, Paul A et al (2018) Identification of MS-specific serum miRNAs in an international multicenter study. Neurol Neuroimmunol Neuroinflammation 5:e491
Muñoz-San Martín M, Reverter G, Robles-Cedeño R et al (2019) Analysis of miRNA signatures in CSF identifies upregulation of miR-21 and miR-146a/b in patients with multiple sclerosis and active lesions. J Neuroinflammation 16:220. https://doi.org/10.1186/s12974-019-1590-5
Regev K, Paul A, Healy B et al (2016) Comprehensive evaluation of serum microRNAs as biomarkers in multiple sclerosis. Neurol Neuroimmunol Neuroinflamm 3:e267. https://doi.org/10.1212/NXI.0000000000000267
Bergman P, Piket E, Khademi M et al (2016) Circulating miR-150 in CSF is a novel candidate biomarker for multiple sclerosis. Neurol Neuroimmunol Neuroinflamm 3:e219. https://doi.org/10.1212/NXI.0000000000000219
Al-Ghezi ZZ, Miranda K, Nagarkatti M, Nagarkatti PS (2019) Combination of Cannabinoids, Δ9- Tetrahydrocannabinol and Cannabidiol, Ameliorates Experimental Multiple Sclerosis by Suppressing Neuroinflammation Through Regulation of miRNA-Mediated Signaling Pathways. Front Immunol 10:1921. https://doi.org/10.3389/fimmu.2019.01921
Criste G, Trapp B, Dutta R (2014) Axonal loss in multiple sclerosis: causes and mechanisms. Handb Clin Neurol 122:101–113. https://doi.org/10.1016/B978-0-444-52001-2.00005-4
Ahn JJ, Abu-Rub M, Miller RH (2021) B Cells in Neuroinflammation: New Perspectives and Mechanistic Insights. Cells 10. https://doi.org/10.3390/cells10071605
Asha MZI, Al-Asaad Y, Khalil SFH (2021) The comparative efficacy and safety of anti-CD20 monoclonal antibodies for relapsing-remitting multiple sclerosis: A network meta-analysis. IBRO Neurosci Rep 11:103–111. https://doi.org/10.1016/j.ibneur.2021.08.003
Lassmann H (2013) Pathology and disease mechanisms in different stages of multiple sclerosis. J Neurol Sci 333:1–4. https://doi.org/10.1016/j.jns.2013.05.010
Romme Christensen J, Komori M, von Essen MR et al (2019) CSF inflammatory biomarkers responsive to treatment in progressive multiple sclerosis capture residual inflammation associated with axonal damage. Mult Scler 25:937–946. https://doi.org/10.1177/1352458518774880
Ghorbani S, Talebi F, Chan WF et al (2017) MicroRNA-181 Variants Regulate T Cell Phenotype in the Context of Autoimmune Neuroinflammation. Front Immunol 8:758. https://doi.org/10.3389/fimmu.2017.00758
Kleiter I, Song J, Lukas D et al (2010) Smad7 in T cells drives T helper 1 responses in multiple sclerosis and experimental autoimmune encephalomyelitis. Brain 133:1067–1081
Galloway DA, Blandford SN, Berry T et al (2019) miR-223 promotes regenerative myeloid cell phenotype and function in the demyelinated central nervous system. Glia 67:857–869. https://doi.org/10.1002/glia.23576
Qin H, Niyongere SA, Lee SJ et al (2008) Expression and functional significance of SOCS-1 and SOCS-3 in astrocytes. J Immunol 181:3167–3176. https://doi.org/10.4049/jimmunol.181.5.3167
Cianciulli A, Calvello R, Porro C et al (2017) Understanding the role of SOCS signaling in neurodegenerative diseases: Current and emerging concepts. Cytokine Growth Factor Rev 37:67–79. https://doi.org/10.1016/j.cytogfr.2017.07.005
Liu X, He F, Pang R et al (2014) Interleukin-17 (IL-17)-induced MicroRNA 873 (miR-873) Contributes to the Pathogenesis of Experimental Autoimmune Encephalomyelitis by Targeting A20 Ubiquitin-editing Enzyme. J Biol Chem 289:28971–28986. https://doi.org/10.1074/jbc.M114.577429
Liu X, Zhou F, Yang Y et al (2019) MiR-409-3p and MiR-1896 co-operatively participate in IL-17-induced inflammatory cytokine production in astrocytes and pathogenesis of EAE mice via targeting SOCS3/STAT3 signaling. Glia 67:101–112. https://doi.org/10.1002/glia.23530
Talebi F, Ghorbani S, Chan WF et al (2017) MicroRNA-142 regulates inflammation and T cell differentiation in an animal model of multiple sclerosis. J Neuroinflammation 14:55. https://doi.org/10.1186/s12974-017-0832-7
Bergman P, James T, Kular L et al (2013) Next-generation sequencing identifies microRNAs that associate with pathogenic autoimmune neuroinflammation in rats. J Immunol 190:4066–4075. https://doi.org/10.4049/jimmunol.1200728
Giunti D, Marini C, Parodi B et al (2019) Exosome-shuttled miRNAs contribute to the modulation of the neuroinflammatory microglia phenotype by mesenchymal stem cells. Implication for amyotrophic lateral sclerosis. Research Square. https://doi.org/10.21203/rs.2.17858/v1
Giunti D, Marini C, Parodi B et al (2021) Role of miRNAs shuttled by mesenchymal stem cell-derived small extracellular vesicles in modulating neuroinflammation. Sci Rep 11:1–17. https://doi.org/10.1038/s41598-021-81039-4
Finardi A, Diceglie M, Carbone L et al (2020) Mir106b-25 and Mir17-92 Are Crucially Involved in the Development of Experimental Neuroinflammation. Front Neurol 11:912. https://doi.org/10.3389/fneur.2020.00912
Hosseiny M, Newsome SD, Yousem DM (2020) Radiologically Isolated Syndrome: A Review for Neuroradiologists. AJNR Am J Neuroradiol 41:1542–1549. https://doi.org/10.3174/ajnr.A6649
Muñoz-San Martín M, Torras S, Robles-Cedeño R et al (2020) Radiologically isolated syndrome: targeting miRNAs as prognostic biomarkers. Epigenomics 12:2065–2076. https://doi.org/10.2217/epi-2020-0172
Perdaens O, Dang HA, D’Auria L, van Pesch V (2020) CSF microRNAs discriminate MS activity and share similarity to other neuroinflammatory disorders. Neurol Neuroimmunol Neuroinflamm 7. https://doi.org/10.1212/NXI.0000000000000673
Hemmer B, Kerschensteiner M, Korn T (2015) Role of the innate and adaptive immune responses in the course of multiple sclerosis. Lancet Neurol 14:406–419. https://doi.org/10.1016/S1474-4422(14)70305-9
Amoruso A, Blonda M, Gironi M et al (2020) Immune and central nervous system-related miRNAs expression profiling in monocytes of multiple sclerosis patients. Sci Rep 10:6125. https://doi.org/10.1038/s41598-020-63282-3
Wu D, Cerutti C, Lopez-Ramirez MA et al (2015) Brain endothelial miR-146a negatively modulates T-cell adhesion through repressing multiple targets to inhibit NF-κB activation. J Cereb Blood Flow Metab 35:412–423. https://doi.org/10.1038/jcbfm.2014.207
Banerjee S, Cui H, Xie N et al (2013) miR-125a-5p regulates differential activation of macrophages and inflammation. J Biol Chem 288:35428–35436. https://doi.org/10.1074/jbc.M112.426866
Essandoh K, Li Y, Huo J, Fan G-C (2016) MiRNA-Mediated Macrophage Polarization and its Potential Role in the Regulation of Inflammatory Response. Shock 46:122–131. https://doi.org/10.1097/SHK.0000000000000604
Moore CS, Rao VTS, Durafourt BA et al (2013) miR-155 as a multiple sclerosis-relevant regulator of myeloid cell polarization. Ann Neurol 74:709–720. https://doi.org/10.1002/ana.23967
Lu L, McCurdy S, Huang S et al (2016) Time Series miRNA-mRNA integrated analysis reveals critical miRNAs and targets in macrophage polarization. Sci Rep 6:37446. https://doi.org/10.1038/srep37446
Xiao M, Xiao ZJ, Yang B et al (2020) Blood-Brain Barrier: More Contributor to Disruption of Central Nervous System Homeostasis Than Victim in Neurological Disorders. Front Neurosci 14:764. https://doi.org/10.3389/fnins.2020.00764
de Vries HE, Kooij G, Frenkel D et al (2012) Inflammatory events at blood-brain barrier in neuroinflammatory and neurodegenerative disorders: implications for clinical disease. Epilepsia 53(Suppl 6):45–52. https://doi.org/10.1111/j.1528-1167.2012.03702.x
Kamphuis WW, Derada Troletti C, Reijerkerk A et al (2015) The blood-brain barrier in multiple sclerosis: microRNAs as key regulators. CNS Neurol Disord Drug Targets 14:157–167. https://doi.org/10.2174/1871527314666150116125246
Cerutti C, Edwards LJ, de Vries HE et al (2017) MiR-126 and miR-126* regulate shear-resistant firm leukocyte adhesion to human brain endothelium. Sci Rep 7:45284. https://doi.org/10.1038/srep45284
Lopez-Ramirez MA, Wu D, Pryce G et al (2014) MicroRNA-155 negatively affects blood-brain barrier function during neuroinflammation. FASEB J 28:2551–2565. https://doi.org/10.1096/fj.13-248880
Louveau A, Herz J, Alme MN et al (2018) CNS lymphatic drainage and neuroinflammation are regulated by meningeal lymphatic vasculature. Nat Neurosci 21:1380–1391. https://doi.org/10.1038/s41593-018-0227-9
Hoye ML, Archambault AS, Gordon TM et al (2018) MicroRNA signature of central nervous system-infiltrating dendritic cells in an animal model of multiple sclerosis. Immunology 155:112–122. https://doi.org/10.1111/imm.12934
Hemond CC, Healy BC, Tauhid S et al (2019) MRI phenotypes in MS: Longitudinal changes and miRNA signatures. Neurol Neuroimmunol Neuroinflamm 6:e530. https://doi.org/10.1212/NXI.0000000000000530
Masoumi F, Ghorbani S, Talebi F et al (2019) Malat1 long noncoding RNA regulates inflammation and leukocyte differentiation in experimental autoimmune encephalomyelitis. J Neuroimmunol 328:50–59. https://doi.org/10.1016/j.jneuroim.2018.11.013
Liu X, Zhou F, Wang W et al (2021) IL-9-triggered lncRNA Gm13568 regulates Notch1 in astrocytes through interaction with CBP/P300: contribute to the pathogenesis of experimental autoimmune encephalomyelitis. J Neuroinflammation 18:108. https://doi.org/10.1186/s12974-021-02156-5
Zhang F, Liu G, Li D et al (2018) DDIT4 and Associated lncDDIT4 Modulate Th17 Differentiation through the DDIT4/TSC/mTOR Pathway. J Immunol 200:1618–1626. https://doi.org/10.4049/jimmunol.1601689
Gupta M, Martens K, Metz LM et al (2019) Long noncoding RNAs associated with phenotypic severity in multiple sclerosis. Mult Scler Relat Disord 36:101407. https://doi.org/10.1016/j.msard.2019.101407
Zhang F, Liu G, Wei C et al (2017) Linc-MAF-4 regulates Th1/Th2 differentiation and is associated with the pathogenesis of multiple sclerosis by targeting MAF. FASEB J 31:519–525. https://doi.org/10.1096/fj.201600838R
Shannon P (2003) Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res 13:2498–2504
Piñero J, Saüch J, Sanz F, Furlong LI (2021) The DisGeNET cytoscape app: Exploring and visualizing disease genomics data. Comput Struct Biotechnol J 19:2960–2967. https://doi.org/10.1016/j.csbj.2021.05.015
Acknowledgements
This work was written by the members of the International and Intercontinental Cardiovascular and Cardiometabolic Research Team (I-COMET; www.icomet.science).
Funding
The work was supported financially as part of the research grant ‘OPUS’ from National Science Centre, Poland (grant number 2018/31/B/NZ7/01137).
Author information
Authors and Affiliations
Contributions
AN, CE, AS, DMG contributed to writing—original draft preparation; CE, AN, ZW, MP, DMG, AC, JP, PS, JJP helped in writing—revision and editing; CE, ZW contributed to visualization; CE, JP, MP, DMG helped in supervision.
Corresponding author
Ethics declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing Interests
Authors declare no conflict of interest related to this work.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Nowak, A., Wicik, Z., Wolska, M. et al. The role of non-coding RNAs in neuroinflammatory process in multiple sclerosis. Mol Neurobiol 59, 4651–4668 (2022). https://doi.org/10.1007/s12035-022-02854-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-022-02854-y