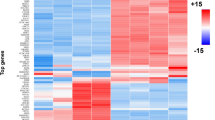
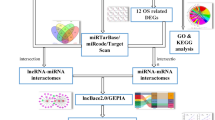
Abstract
Transcription factor (TF) and microRNA (miRNA) are two best characterized gene regulators that have been found to play an important role in gene regulation. However, high throughput screening the interaction relationships between transcription factors, microRNAs, and target genes in gliomas remains rare. Using GSE16666 and GSE13091 datasets downloaded from Gene Expression Omnibus data, we first screened the differentially expressed genes in gliomas. We explored the regulation relationship among TFs, miRNAs and target genes by different algorithms. The underlying molecular mechanisms of these crucial target genes were investigated by Gene Ontology function and Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis. Our study has developed three regulation relationships between two TFs and three miRNAs, including TP53/hsa-mir-155, TP53/hsa-mir-125b, and KLF2/hsa-mir-126. In addition, we also constructed a regulation network of the target genes by transcription factors and miRNAs. Some of them had been demonstrated to be involved in glioma progression via various pathways. For example, ATP2B2 target gene could be regulated by has-mir-181a to involve in calcium signaling pathway. RB1 could be regulated by has-miR-26a to participate in pathways in cancer. Smad7 could be regulated by has-miR-21 via intracellular TGF-β signal transduction. We constructed a comprehensive regulatory network which was found to play an important role in gliomas progression.




Similar content being viewed by others
References
Yamanaka R (2008) Cell-and peptide-based immunotherapeutic approaches for glioma. Trends mol med 14(5):228–235
Chen K, Rajewsky N (2007) The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet 8(2):93–103
Zhou Q, Wong WH (2004) CisModule: de novo discovery of cis-regulatory modules by hierarchical mixture modeling. Proc Natl Acad Sci USA 101(33):12114
Yan S, Berquin IM, Troen BR, Sloane BF (2000) Transcription of human cathepsin B is mediated by Sp1 and Ets family factors in glioma. DNA Cell Biol 19(2):79–91
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116(2):281–297
Chan JA, Krichevsky AM, Kosik KS (2005) MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res 65(14):6029
Gabriely G, Wurdinger T, Kesari S, Esau CC, Burchard J, Linsley PS, Krichevsky AM (2008) MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol Cell Biol 28(17):5369–5380
Sasayama T, Nishihara M, Kondoh T, Hosoda K, Kohmura E (2009) MicroRNA-10b is overexpressed in malignant glioma and associated with tumor invasive factors, uPAR and RhoC. Int J Cancer 125(6):1407–1413
Kefas B, Godlewski J, Comeau L, Li Y, Abounader R, Hawkinson M, Lee J, Fine H, Chiocca EA, Lawler S (2008) microRNA-7 inhibits the epidermal growth factor receptor and the Akt pathway and is down-regulated in glioblastoma. Cancer Res 68(10):3566
Su N, Wang Y, Qian M, Deng M (2010) Combinatorial regulation of transcription factors and microRNAs. BMC Syst Biol 4(1):150
Shalgi R, Lieber D, Oren M, Pilpel Y (2007) Global and local architecture of the mammalian microRNA–transcription factor regulatory network. PLoS Comput Biol 3(7):e131
Weiner A, Hughes A, Yassour M, Rando OJ, Friedman N (2010) High-resolution nucleosome mapping reveals transcription-dependent promoter packaging. Genome Res 20(1):90–100
Wurdinger T, Tannous BA, Saydam O, Skog J, Grau S, Soutschek J, Weissleder R, Breakefield XO, Krichevsky AM (2008) miR-296 regulates growth factor receptor overexpression in angiogenic endothelial cells. Cancer Cell 14(5):382–393
Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV et al (2003) TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res 31(1):374–378
John B, Sander C, Marks DS (2006) Prediction of human microRNA targets. Methods Mol Biol 342:101–113
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc Ser B (Methodological):289–300
Margolin AA, Nemenman I, Basso K, Wiggins C, Stolovitzky G, Dalla Favera R, Califano A (2006) ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics 7(Suppl 1):S7
Wang J, Lu M, Qiu C, Cui Q (2010) TransmiR: a transcription factor-microRNA regulation database. Nucleic Acids Res 38(Database issue):D119-122
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT et al (2000) Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat Genet 25(1):25–29
Kanehisa M (2002) The KEGG database. Novartis Found Symp 247:91–101 discussion 101–103, 119–128, 244–152
Suh SS, Yoo JY, Nuovo GJ, Jeon YJ, Kim S, Lee TJ, Kim T, Bakàcs A, Alder H, Kaur B (2012) MicroRNAs/TP53 feedback circuitry in glioblastoma multiforme. Proc Natl Acad Sci 109(14):5316–5321
D’Urso PI, D’Urso OF, Storelli C, Mallardo M, Gianfreda CD, Montinaro A, Cimmino A, Pietro C, Marsigliante S (2012) miR-155 is up-regulated in primary and secondary glioblastoma and promotes tumour growth by inhibiting GABA receptors. Int j oncol 41(1):228–234
Cobbs CS, Whisenhunt TR, Wesemann DR, Harkins LE, Van Meir EG, Samanta M (2003) Inactivation of wild-type p53 protein function by reactive oxygen and nitrogen species in malignant glioma cells. Cancer Res 63(24):8670
Gong H, Liu CM, Liu DP, Liang CC (2005) The role of small RNAs in human diseases: potential troublemaker and therapeutic tools. Med Res Rev 25(3):361–381
Xia HF, He TZ, Liu CM, Cui Y, Song PP, Jin XH, Ma X (2009) MiR-125b expression affects the proliferation and apoptosis of human glioma cells by targeting Bmf. Cell Physiol Biochem 23(4–6):347–358
Le MTN, Teh C, Shyh-Chang N, Xie H, Zhou B, Korzh V, Lodish HF, Lim B (2009) MicroRNA-125b is a novel negative regulator of p53. Genes Dev 23(7):862–876
Feng J, Kim ST, Liu W, Kim JW, Zhang Z, Zhu Y, Berens M, Sun J, Xu J (2012) An integrated analysis of germline and somatic, genetic and epigenetic alterations at 9p21. 3 in glioblastoma. Cancer 118(1):232–240
Harris TA, Yamakuchi M, Kondo M, Oettgen P, Lowenstein CJ (2010) Ets-1 and Ets-2 regulate the expression of microRNA-126 in endothelial cells. Arterioscler Thromb Vasc Biol 30(10):1990–1997
Monteith GR, McAndrew D, Faddy HM, Roberts-Thomson SJ (2007) Calcium and cancer: targeting Ca2+transport. Nat Rev Cancer 7(7):519–530
Kovacs GG, Zsembery A, Anderson SJ, Komlosi P, Gillespie GY, Bell PD, Benos DJ, Fuller CM (2005) Changes in intracellular Ca2+ and pH in response to thapsigargin in human glioblastoma cells and normal astrocytes. Am J Physiol Cell Physiol 289(2):C361–C371
Chen YJ, Lin JK, Lin-Shiau SY (1999) Proliferation arrest and induction of CDK inhibitors p21 and p27 by depleting the calcium store in cultured C6 glioma cells. Eur J Cell Biol 78(11):824–831
Guimaraes-Sternberg C, Meerson A, Shaked I, Soreq H (2006) MicroRNA modulation of megakaryoblast fate involves cholinergic signaling. Leuk Res 30(5):583–595
Harbour JW, Dean DC (2000) Rb function in cell-cycle regulation and apoptosis. Nat Cell Biol 2(4):E65–E67
Nakamura M, Yonekawa Y, Kleihues P, Ohgaki H (2001) Promoter hypermethylation of the RB1 gene in glioblastomas. Lab Invest 81(1):77–82
Mathivanan J, Rohini K, Gope ML, Anandh B, Gope R (2007) Altered structure and deregulated expression of the tumor suppressor gene retinoblastoma (RB1) in human brain tumors. Mol Cell Biochem 302(1):67–77
Kim H, Huang W, Jiang X, Pennicooke B, Park PJ, Johnson MD (2010) Integrative genome analysis reveals an oncomir/oncogene cluster regulating glioblastoma survivorship. Proc Natl Acad Sci 107(5):2183–2188
Seoane J, Le HV, Shen L, Anderson SA, Massagué J (2004) Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell 117(2):211–223
Piek E, Westermark U, Kastemar M, Heldin CH, van Zoelen E, Nistér M, Ten Dijke P (1999) Expression of transforming-growth-factor (TGF)-β receptors and Smad proteins in glioblastoma cell lines with distinct responses to TGF-β1. Int J Cancer 80(5):756–763
Papagiannakopoulos T, Shapiro A, Kosik KS (2008) MicroRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Cancer Res 68(19):8164
Liu G, Friggeri A, Yang Y, Milosevic J, Ding Q, Thannickal VJ, Kaminski N, Abraham E (2010) miR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J Exp Med 207(8):1589–1597
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yu, J., Cai, X., He, J. et al. Microarray-based analysis of gene regulation by transcription factors and microRNAs in glioma. Neurol Sci 34, 1283–1289 (2013). https://doi.org/10.1007/s10072-012-1228-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10072-012-1228-1