Abstract
Introduction
Dermatomyositis (DM) is a rare inflammatory disease characterized by the invasion of the skin and muscles. Environmental, genetic, and immunological factors contribute to disease pathology. To date, no bioinformatics studies have been conducted on the potential pathogenic genes and immune cell infiltration in DM. Therefore, we aimed to identify differentially expressed genes (DEGs) and immune cells, as well as potential pathogenic genes and immune characteristics, which may be useful for the diagnosis and treatment of DM.
Method
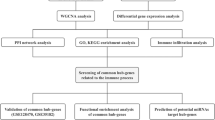
GSE1551, GSE5370, GSE39454, and GSE48280 from Gene Expression Omnibus were included in our study. Limma, ClusterProfiler, and Kyoto Encyclopedia of Genes and Genomes were used to identify DEGs, Gene Ontology (GO), and perform pathway analyses, respectively. Cytoscape was used to construct the protein-protein interaction (PPI) network. Small-molecule drugs were identified using a connectivity map (CMap), and the TIMER database was used to identify infiltrating cells.
Results
DEG analysis identified 12 downregulated and 163 upregulated genes. GO analysis showed that DEGs were enriched in immune-related pathways. Ten hub genes were identified from the PPI network. Additionally, CMap analysis showed that caffeic acid, sulfaphenazole, molindone, tiabendazole, and bacitracin were potential small-molecule drugs with therapeutic significance. We identified eight immune cells with differential infiltration in patients with DM and controls. Finally, we constructed a powerful diagnostic model based on memory B cells, M1, and M2 macrophages.
Conclusions
This study explored the potential molecular mechanism and immunological landscape of DM and may guide future research and treatment of DM.
Key Points
• We explored the molecular mechanism and immunological landscape of dermatomyositis.
• GO analysis showed that DEGs were enriched in immune-related pathways.
• We predicted small-molecular drugs with potential therapeutic significance based on bioanalytical techniques.
• We identified six immune cells with differential infiltration in patients with DM and controls.





Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in this published article.
References
Qudsiya Z, Waseem M (2020) Dermatomyositis. In: StatPearls. StatPearls Publishing Copyright © 2020, StatPearls Publishing LLC., Treasure Island (FL)
Dalakas MC, Hohlfeld R (2003) Polymyositis and dermatomyositis. Lancet (London, England) 362(9388):971–982. https://doi.org/10.1016/s0140-6736(03)14368-1
Tournadre A, Miossec P (2013) A critical role for immature muscle precursors in myositis. Nat Rev Rheumatol 9(7):438–442. https://doi.org/10.1038/nrrheum.2013.26
DeWane ME, Waldman R, Lu J (2020) Dermatomyositis: clinical features and pathogenesis. J Am Acad Dermatol 82(2):267–281. https://doi.org/10.1016/j.jaad.2019.06.1309
Shao C, Li S, Sun Y, Zhang Y, Xu K, Zhang X, Huang H (2020) Clinical characteristics and prognostic analysis of Chinese dermatomyositis patients with malignancies. Medicine 99(34):e21899. https://doi.org/10.1097/md.0000000000021899
Adler BL, Christopher-Stine L (2018) Triggers of inflammatory myopathy: insights into pathogenesis. Discov Med 25(136):75–83
Dourmishev AL, Dourmishev LA (1999) Dermatomyositis and drugs. Adv Exp Med Biol 455:187–191. https://doi.org/10.1007/978-1-4615-4857-7_27
O'Hanlon TP, Carrick DM, Arnett FC, Reveille JD, Carrington M, Gao X, Oddis CV, Morel PA, Malley JD, Malley K, Dreyfuss J, Shamim EA, Rider LG, Chanock SJ, Foster CB, Bunch T, Plotz PH, Love LA, Miller FW (2005) Immunogenetic risk and protective factors for the idiopathic inflammatory myopathies: distinct HLA-A, -B, -Cw, -DRB1 and -DQA1 allelic profiles and motifs define clinicopathologic groups in caucasians. Medicine 84(6):338–349. https://doi.org/10.1097/01.md.0000189818.63141.8c
O'Hanlon TP, Rider LG, Mamyrova G, Targoff IN, Arnett FC, Reveille JD, Carrington M, Gao X, Oddis CV, Morel PA, Malley JD, Malley K, Shamim EA, Chanock SJ, Foster CB, Bunch T, Reed AM, Love LA, Miller FW (2006) HLA polymorphisms in African Americans with idiopathic inflammatory myopathy: allelic profiles distinguish patients with different clinical phenotypes and myositis autoantibodies. Arthritis Rheum 54(11):3670–3681. https://doi.org/10.1002/art.22205
Gao X, Han L, Yuan L, Yang Y, Gou G, Sun H, Lu L, Bao L (2014) HLA class II alleles may influence susceptibility to adult dermatomyositis and polymyositis in a Han Chinese population. BMC Dermatol 14:9. https://doi.org/10.1186/1471-5945-14-9
Lahouti AH, Christopher-Stine L (2015) Polymyositis and dermatomyositis: novel insights into the pathogenesis and potential therapeutic targets. Discov Med 19(107):463–470
Greenberg SA (2007) A gene expression approach to study perturbed pathways in myositis. Curr Opin Rheumatol 19(6):536–541. https://doi.org/10.1097/BOR.0b013e3282efe261
Schultz HY, Dutz JP, Furukawa F, Goodfield MJ, Kuhn A, Lee LA, Nyberg F, Szepietowski JC, Sontheimer RD, Werth VP (2015) From pathogenesis, epidemiology, and genetics to definitions, diagnosis, and treatments of cutaneous lupus erythematosus and dermatomyositis: a report from the 3rd International Conference on Cutaneous Lupus Erythematosus (ICCLE) 2013. The Journal of investigative dermatology 135(1):7–12. https://doi.org/10.1038/jid.2014.316
Ghirardello A, Zampieri S, Tarricone E, Iaccarino L, Gorza L, Doria A (2011) Cutting edge issues in polymyositis. Clin Rev Allergy Immunol 41(2):179–189. https://doi.org/10.1007/s12016-010-8238-7
Petryszak R, Burdett T, Fiorelli B, Fonseca NA, Gonzalez-Porta M, Hastings E, Huber W, Jupp S, Keays M, Kryvych N, McMurry J, Marioni JC, Malone J, Megy K, Rustici G, Tang AY, Taubert J, Williams E, Mannion O, Parkinson HE, Brazma A (2014) Expression atlas update--a database of gene and transcript expression from microarray- and sequencing-based functional genomics experiments. Nucleic Acids Res 42(Database issue):D926–D932. https://doi.org/10.1093/nar/gkt1270
Greenberg SA, Pinkus JL, Pinkus GS, Burleson T, Sanoudou D, Tawil R, Barohn RJ, Saperstein DS, Briemberg HR, Ericsson M, Park P, Amato AA (2005) Interferon-alpha/beta-mediated innate immune mechanisms in dermatomyositis. Ann Neurol 57(5):664–678. https://doi.org/10.1002/ana.20464
Zhu W, Streicher K, Shen N, Higgs BW, Morehouse C, Greenlees L, Amato AA, Ranade K, Richman L, Fiorentino D, Jallal B, Greenberg SA, Yao Y (2012) Genomic signatures characterize leukocyte infiltration in myositis muscles. BMC Med Genet 5:53. https://doi.org/10.1186/1755-8794-5-53
Suárez-Calvet X, Gallardo E, Nogales-Gadea G, Querol L, Navas M, Díaz-Manera J, Rojas-Garcia R, Illa I (2014) Altered RIG-I/DDX58-mediated innate immunity in dermatomyositis. J Pathol 233(3):258–268. https://doi.org/10.1002/path.4346
Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD (2012) The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics (Oxford, England) 28(6):882–883. https://doi.org/10.1093/bioinformatics/bts034
Subramanian A, Narayan R, Corsello SM, Peck DD, Natoli TE, Lu X, Gould J, Davis JF, Tubelli AA, Asiedu JK, Lahr DL, Hirschman JE, Liu Z, Donahue M, Julian B, Khan M, Wadden D, Smith IC, Lam D, Liberzon A, Toder C, Bagul M, Orzechowski M, Enache OM, Piccioni F, Johnson SA, Lyons NJ, Berger AH, Shamji AF, Brooks AN, Vrcic A, Flynn C, Rosains J, Takeda DY, Hu R, Davison D, Lamb J, Ardlie K, Hogstrom L, Greenside P, Gray NS, Clemons PA, Silver S, Wu X, Zhao WN, Read-Button W, Wu X, Haggarty SJ, Ronco LV, Boehm JS, Schreiber SL, Doench JG, Bittker JA, Root DE, Wong B, Golub TR (2017) A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell 171 (6):1437-1452.e1417. doi:https://doi.org/10.1016/j.cell.2017.10.049
Yu G, Wang LG, Han Y, He QY (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. Omics 16(5):284–287. https://doi.org/10.1089/omi.2011.0118
Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, Jensen LJ, von Mering C (2017) The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible 45 (D1):D362-d368. doi:https://doi.org/10.1093/nar/gkw937
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13(11):2498–2504. https://doi.org/10.1101/gr.1239303
Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, Li B, Liu XS (2020) TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res 48(W1):W509–w514. https://doi.org/10.1093/nar/gkaa407
Plattner C, Finotello F, Rieder D (2020) Deconvoluting tumor-infiltrating immune cells from RNA-seq data using quanTIseq. Methods Enzymol 636:261–285. https://doi.org/10.1016/bs.mie.2019.05.056
Aran D, Hu Z, Butte AJ (2017) xCell: digitally portraying the tissue cellular heterogeneity landscape. 18 (1):220. doi:https://doi.org/10.1186/s13059-017-1349-1
Collins DM, Madden SF, Gaynor N, AlSultan D, Le Gal M, Eustace AJ (2020) Effects of HER family-targeting tyrosine kinase inhibitors on antibody-dependent cell-mediated cytotoxicity in HER2-expressing breast. Cancer. doi:https://doi.org/10.1158/1078-0432.ccr-20-2007
Racle J, de Jonge K, Baumgaertner P, Speiser DE, Gfeller D (2017) Simultaneous enumeration of cancer and immune cell types from bulk tumor gene expression data 6. doi:https://doi.org/10.7554/eLife.26476
Li W, Zhang Z, Wang ZM (2020) Differential immune cell infiltrations between healthy periodontal and chronic periodontitis tissues. BMC oral health 20(1):293. https://doi.org/10.1186/s12903-020-01287-0
Xin Y, Zhang S, Deng Z, Zeng D, Li J, Zhang Y (2020) Identification and verification immune-related regulatory network in acne. International immunopharmacology 89 (Pt B):107083. doi:https://doi.org/10.1016/j.intimp.2020.107083
Xiu MX, Liu YM, Chen GY, Hu C, Kuang BH (2020) Identifying hub genes, key pathways and immune cell infiltration characteristics in pediatric and adult ulcerative colitis by integrated bioinformatic analysis. Digestive diseases and sciences. doi:https://doi.org/10.1007/s10620-020-06611-w
Ren C, Li M, Du W, Lü J, Zheng Y, Xu H, Quan R (2020) Comprehensive bioinformatics analysis reveals hub genes and inflammation state of rheumatoid arthritis. 2020:6943103. doi:https://doi.org/10.1155/2020/6943103
Cao Y, Tang W, Tang W (2019) Immune cell infiltration characteristics and related core genes in lupus nephritis: results from bioinformatic analysis. 20 (1):37. doi:https://doi.org/10.1186/s12865-019-0316-x
Newman AM, Liu CL, Green MR (2015) Robust enumeration of cell subsets from tissue expression profiles. 12 (5):453-457. doi:https://doi.org/10.1038/nmeth.3337
Moneta GM, Pires Marafon D, Marasco E (2019) Muscle expression of type I and type II interferons is increased in juvenile dermatomyositis and related to clinical and histologic features 71 (6):1011–1021. doi:https://doi.org/10.1002/art.40800
Peng QL, Lin JM, Zhang YB, Zhang XZ, Wang PP, Wu TT, Yu J, Dong XQ, Gu ML, Wang GC (2018) Targeted capture sequencing identifies novel genetic variations in Chinese patients with idiopathic inflammatory myopathies. Int J Rheum Dis 21(8):1619–1626. https://doi.org/10.1111/1756-185x.13350
Rothwell S, Cooper RG, Lundberg IE, Miller FW, Gregersen PK, Bowes J, Vencovsky J, Danko K, Limaye V, Selva-O'Callaghan A, Hanna MG, Machado PM, Pachman LM, Reed AM, Rider LG, Cobb J, Platt H, Molberg Ø, Benveniste O, Mathiesen P, Radstake T, Doria A, De Bleecker J, De Paepe B, Maurer B, Ollier WE, Padyukov L, O'Hanlon TP, Lee A, Amos CI, Gieger C, Meitinger T, Winkelmann J, Wedderburn LR, Chinoy H, Lamb JA (2016) Dense genotyping of immune-related loci in idiopathic inflammatory myopathies confirms HLA alleles as the strongest genetic risk factor and suggests different genetic background for major clinical subgroups. Ann Rheum Dis 75(8):1558–1566. https://doi.org/10.1136/annrheumdis-2015-208119
Furuya T, Hakoda M, Higami K, Ueda H, Tsuchiya N, Tokunaga K, Kamatani N, Kashiwazaki S (1998) Association of HLA class I and class II alleles with myositis in Japanese patients. J Rheumatol 25(6):1109–1114
Tournadre A, Lenief V, Eljaafari A, Miossec P (2012) Immature muscle precursors are a source of interferon-β in myositis: role of Toll-like receptor 3 activation and contribution to HLA class I up-regulation. Arthritis Rheum 64(2):533–541. https://doi.org/10.1002/art.33350
Franzi S, Salajegheh M, Nazareno R, Greenberg SA (2013) Type 1 interferons inhibit myotube formation independently of upregulation of interferon-stimulated gene 15. PLoS One 8(6):e65362. https://doi.org/10.1371/journal.pone.0065362
Bilgic H, Ytterberg SR, Amin S, McNallan KT, Wilson JC, Koeuth T, Ellingson S, Newman B, Bauer JW, Peterson EJ, Baechler EC, Reed AM (2009) Interleukin-6 and type I interferon-regulated genes and chemokines mark disease activity in dermatomyositis. Arthritis Rheum 60(11):3436–3446. https://doi.org/10.1002/art.24936
Greenberg SA, Higgs BW, Morehouse C, Walsh RJ, Kong SW, Brohawn P, Zhu W, Amato A, Salajegheh M, White B, Kiener PA, Jallal B, Yao Y (2012) Relationship between disease activity and type 1 interferon- and other cytokine-inducible gene expression in blood in dermatomyositis and polymyositis. Genes Immun 13(3):207–213. https://doi.org/10.1038/gene.2011.61
Aouba A, Georgin-Lavialle S, Terrier B, Guillevin L, Authier FJ (2011) Anti-PL7 antisynthetase syndrome under interferon therapy. Joint bone spine 78(1):94–97. https://doi.org/10.1016/j.jbspin.2010.07.012
Ladislau L, Suárez-Calvet X, Toquet S, Landon-Cardinal O, Amelin D, Depp M, Rodero MP, Hathazi D, Duffy D, Bondet V, Preusse C, Bienvenu B, Rozenberg F, Roos A, Benjamim CF, Gallardo E, Illa I, Mouly V, Stenzel W, Butler-Browne G, Benveniste O, Allenbach Y (2018) JAK inhibitor improves type I interferon induced damage: proof of concept in dermatomyositis. Brain : a journal of neurology 141(6):1609–1621. https://doi.org/10.1093/brain/awy105
Hornung T, Wenzel J (2014) Innate immune-response mechanisms in dermatomyositis: an update on pathogenesis, diagnosis and treatment. Drugs 74(9):981–998. https://doi.org/10.1007/s40265-014-0240-6
Kaufmann J, Hunzelmann N, Genth E, Krieg T (2005) The clinical spectrum of dermatomyositis. Journal der Deutschen Dermatologischen Gesellschaft = Journal of the German Society of Dermatology : JDDG 3(3):181–194. https://doi.org/10.1111/j.1610-0378.2005.05006.x
Kee SJ, Kim TJ, Lee SJ, Cho YN, Park SC, Kim JS, Kim JC, Kang HS, Lee SS, Park YW (2009) Dermatomyositis associated with hepatitis B virus-related hepatocellular carcinoma. Rheumatol Int 29(5):595–599. https://doi.org/10.1007/s00296-008-0718-1
Hoesly FJ, Sluzevich JC (2014) Chronic cutaneous varicella zoster virus infection complicating dermatomyositis. J Dermatol 41(4):334–336. https://doi.org/10.1111/1346-8138.12402
Thompson C, Piguet V, Choy E (2018) The pathogenesis of dermatomyositis. Br J Dermatol 179(6):1256–1262. https://doi.org/10.1111/bjd.15607
Wang D, Lei L (2020) Interleukin-35 regulates the balance of Th17 and Treg responses during the pathogenesis of connective tissue diseases. Int J Rheum Dis. https://doi.org/10.1111/1756-185x.13962
Waschbisch A, Schwab N, Ruck T, Stenner MP, Wiendl H (2010) FOXP3+ T regulatory cells in idiopathic inflammatory myopathies. J Neuroimmunol 225(1–2):137–142. https://doi.org/10.1016/j.jneuroim.2010.03.013
Peng QL, Zhang YL, Shu XM, Yang HB, Zhang L, Chen F, Lu X, Wang GC (2015) Elevated serum levels of soluble CD163 in polymyositis and dermatomyositis: associated with macrophage infiltration in muscle tissue. J Rheumatol 42(6):979–987. https://doi.org/10.3899/jrheum.141307
Ragusa F (2019) Dermatomyositis and MIG. La Clinica terapeutica 170(2):e142–e147. https://doi.org/10.7417/ct.2019.2124
Zhou Y, Wang J, Chang Y, Li R, Sun X, Peng L, Zheng W (2020) Caffeic acid phenethyl ester protects against experimental autoimmune encephalomyelitis by regulating T cell activities. 2020:7274342. doi:https://doi.org/10.1155/2020/7274342
Choi JH, Roh KH, Oh H, Park SJ, Ha SM, Kang MS, Lee JH, Jung SY, Song H, Yang JW, Park S (2015) Caffeic acid phenethyl ester lessens disease symptoms in an experimental autoimmune uveoretinitis mouse model. Exp Eye Res 134:53–62. https://doi.org/10.1016/j.exer.2015.03.014
Huang C, Liu W, Perry CN, Yitzhaki S, Lee Y, Yuan H, Tsukada YT, Hamacher-Brady A, Mentzer RM Jr, Gottlieb RA (2010) Autophagy and protein kinase C are required for cardioprotection by sulfaphenazole. Am J Phys Heart Circ Phys 298(2):H570–H579. https://doi.org/10.1152/ajpheart.00716.2009
Goktas MT, Karaca RO, Kalkisim S, Cevik L, Kilic L, Akdogan A, Babaoglu MO, Bozkurt A, Bertilsson L, Yasar U (2017) Decreased activity and genetic polymorphisms of CYP2C19 in Behçet’s disease. Basic & clinical pharmacology & toxicology 121(4):266–271. https://doi.org/10.1111/bcpt.12710
Waugaman RM (2009) Potential lower efficacy of molindone among first-generation antipsychotics. Am J Psychiatry 166 (4):491; author reply 492-493. doi:https://doi.org/10.1176/appi.ajp.2009.08111696
Elgebaly SA, Forouhar F, Dore-Duffy P (1984) Thiabendazole-induced suppression of renal damage in a murine model of autoimmune disease. Am J Pathol 115(2):204–211
Matsushima S, Yoshitoshi T, Shichi H (1990) Immunosuppression by gramicidin S of experimental autoimmune uveoretinitis, pinealitis and autoimmune encephalomyelitis. J Ocul Pharmacol 6(3):219–226. https://doi.org/10.1089/jop.1990.6.219
Author information
Authors and Affiliations
Contributions
Conceptualization: Ruxue Yin, Gangjian Wang, and Shengyun Liu; data curation: Ruxue Yin and Lei Zhang; formal analysis: Ruxue Yin, Gangjian Wang, and Shengyun Liu; methodology: Ruxue Yin, Gangjian Wang, and Tianfang Li; resources: Gangjian Wang, Lei Zhang, and Tianfang Li; supervision: Ruxue Yin, Tianfang Li, and Shengyun Liu; writing—original draft: Ruxue Yin and Shengyun Liu; writing—review and editing: Ruxue Yin, Gangjian Wang, and Shengyun Liu.
Corresponding authors
Ethics declarations
Disclosures
None.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Yin, R., Wang, G., Zhang, L. et al. Dermatomyositis: immunological landscape, biomarkers, and potential candidate drugs. Clin Rheumatol 40, 2301–2310 (2021). https://doi.org/10.1007/s10067-020-05568-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10067-020-05568-5