Abstract
Axenfeld–Rieger syndrome (ARS) is a disorder affecting the anterior segment of the eye, often leading to secondary glaucoma and several systemic malformations. It is inherited in an autosomal dominant fashion that has been associated with genetic defects in PITX2 and FOXC1. Known genes CYP1b1, PITX2, and FOXC1 were excluded by Sanger sequencing. The purpose of current study is to identify the underlying genetic causes in ARS family by whole exome sequencing (WES). WES was performed for affected proband of family, and variants were prioritized based on in silico analyses. Segregation analysis of candidate variants was performed in family members. A novel heterozygous PRDM5 missense variant (c.877A>G; p.Lys293Glu) was found to segregate with the disease in an autosomal dominant fashion. The novel missense variant was absent from population-matched controls, the Exome Variant Server, and an in-house exome variant database. The Lys293Glu variant is predicted to be pathogenic and affects a lysine residue that is conserved in different species. Variants in the PRDM5 gene were previously identified in anterior segment defects, i.e., autosomal recessive brittle cornea syndrome and keratoconus. The results of this study suggest that genetic variants in PRDM5 can lead to various syndromic and nonsyndromic disorders affecting the anterior segment of the eye.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Axenfeld–Rieger Syndrome (ARS; OMIM 180500) is a rare developmental disorder inherited in an autosomal dominant manner with an incidence of 1:200,000. ARS is a part of phenotypically heterogeneous group of conditions involving anterior segment dysgenesis, constituting a wide spectrum of developmental anomalies that may affect the cornea, iris, lens, and angle [1]. ARS is characterized by a broad range of abnormalities, with evident ocular and systemic manifestations.
Ocular features observed in ARS patients include iris stromal hypoplasia, polycoria, corectopia, iridogoniodysgenesis, posterior embryotoxon, and iris strands bridging the iridocorneal angle to the trabecular meshwork [2]. Increased ocular pressure (IOP) leading to glaucoma is the major consequence of the anterior segment dysgenesis observed in ARS, with approximately half of the patients developing secondary glaucoma [3]. In addition, ARS patients sometimes present with various systemic abnormalities including facial dysmorphisms (e.g., hypertelorism, telecanthus, maxillary hypoplasia with flattening of the mid-face, prominent forehead, and a broad, flat nasal bridge) and dental abnormalities (e.g., microdontia or hypodontia). In the abdominal region, a failure of involution of the skin results in redundant periumbilical skin. Hypospadia in males, anal stenosis, pituitary abnormalities, and growth retardation may also be observed. Systemic changes other than these are usually not considered as the classical features of ARS [4].
ARS has been associated with mutations in the pituitary homeobox 2 (PITX2; OMIM 601542) gene at 4q25 [5], and the forkhead box C1 (FOXC1; OMIM 601090) gene at 6p25 [6, 7]. A third locus was suggested on 13q14, but a disease-causing gene has not yet been identified [8, 9]. In two isolated ARS cases, deletion of the 16q23-q24 region [10] and deletion of the PAX6 gene at 11p13 [11] have been reported.
The goal of this study was to identify the underlying genetic cause in an autosomal dominant ARS family by whole exome sequencing (WES).
Methods
Clinical evaluation
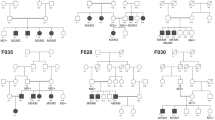
A Pakistani family of Punjabi origin with four affected individuals was included in the study (Fig. 1). The study adhered to the principles of the declaration of Helsinki and was approved by the institutional ethical review board of Al-Shifa Eye Trust Hospital in Rawalpindi. Blood samples were drawn from affected and unaffected family members, after obtaining written informed consent. DNA was extracted using a standard phenol-chloroform method [12].
Segregation of a PRDM5 missense variant in a family with Axenfeld–Rieger syndrome. The c.877A>G; p.Lys293Glu variant is indicated with an M, and the wild-type allele with a +. All affected individuals carry the variant heterozygously, while the unaffected individuals do not carry the variant. The proband is indicated with an arrow
Clinical characterization of affected individuals included funduscopy, and measurement of intraocular pressure (IOP) with Goldmann applanation tonometry. Assessment of visual field defects was performed with a Humphrey Visual Field Analyzer (Carl Zeiss Humphrey Systems, Dublin, CA, USA). Anterior insertion of the iris into the trabecular meshwork with prominent iris processes was observed by gonioscopic examination. All participants were examined by a general physician for the presence or absence of systemic abnormalities.
Sanger sequencing
Known genes cyp1b1, Pitx2, and Foxc1 were excluded by direct Sanger sequencing of the coding exonic and flanking intronic regions of the respective genes. Conditions used to amplify these genes can be provided on request.
Exome sequencing and analysis
To identify the underlying genetic cause of the disease in this family, WES was performed using genomic DNA of the proband (VI:3). Enrichment of exonic sequences was achieved by using the SureSelectXT Human All Exon V.2 Kit (50 Mb) (Agilent Technologies, Inc., Santa Clara, CA, USA). Sequencing was performed on a SOLiD 4 sequencing platform (Life Technologies, Carlsbad, CA, USA). Hg19 reference genome was aligned with the reads obtained using SOLiD LifeScope software V.2.1 (Life Technologies).
To evaluate the pathogenicity of the variants obtained from WES, bioinformatic analysis was performed using PhyloP (nucleotide conservation in various species), Grantham score (amino acid conservation), Sorting Intolerant from Tolerant (SIFT), MutationTaster, and PolyPhen2 (http://genetics.bwh.harvard.edu/pph2/). The presence of potential pathogenic variants was confirmed, and segregation was checked by PCR and Sanger sequencing. Sequencing was performed using the Big Dye Terminator Cycle Sequencing-Ready Reaction Kit (Applied Biosystems) on a 3130 DNA automated sequencer (Applied Biosystems, Foster City, CA, USA) using standard protocols.
Protein conservation
PRDM5 protein sequences from different species were aligned to study the evolutionary conservation of the mutated amino acid Lys293 using Vector NTI Advance 2011.
Protein structure prediction
Homology-based modeling using HOPE [13] was performed to assess the possible structural changes in the mutant protein. The human PDRM5 protein sequence (NM_018699, UniProt ID: Q9NQX1) was used to predict the wild-type and mutant protein structure.
Results
Clinical characterization
The proband (VI:3) has an affected father (V:2) and aunt (V:1), and one affected sister (VI:5) (Fig. 1). The proband was diagnosed at age 4 years. Ocular abnormalities included bilateral buphthalmos, correctopia, iris atrophy, corneal opacity, embryotoxon, posterior subcapsular cataract, and mild vitreous condensation. Funduscopy showed glaucomatous atrophy of the optic nerve with a cup-disc ratio (CDR) of 0.7 and 0.6 for the right and left eyes, respectively, and IOP measured by Goldmann applanation tonometry was 32 mmHg in the right eye and 36 mmHg for the left eye. The proband underwent several surgeries for the right eye, including trabeculectomy with mitomycin C (MMC) when he was 6 years old. For the left eye, cryopexy was done. At 7 years of age, cataract surgery was performed in the right eye, followed by parsplana viterectomy with silicone oil as he developed retinal detachment after trauma. He has typical facial features of ARS such as telecanthus, a broad nasal bridge, micrognathia, and microdontia. His abdominal features included redundant periumbilical skin. Facial dysmorphism in individuals VI:3 and VI:5 included flattening of the mid-face, a broad forehead, a broad nasal bridge, a thin upper lip with a long philtrum, a protruding lower lip and a receding chin (Fig. 2a–f). Both individuals V:1 and V:2 have hearing defects and hip joint anomalies. None of the affected individuals have cardiovascular defects.
Ocular and systemic characteristics of the family with Axenfeld–Rieger syndrome. a Eyes of the proband (VI:3) presented bilateral buphthalmos, corectopia, polycoria, corneal edema, posterior embryotoxon, posterior subcapsular cataract, vascularized corneal opacity, and iris atropy patches in the left eye. b Both eyes of individual (VI:5) showed megalocornea and posterior embryotoxon. The right eye shows polycoria, and the left eye shows corectopia. c Facial dysmorphism in individuals VI:3 included flattening of the mid-face, a broad forehead, a broad nasal bridge, a thin upper lip with a long philtrum, a protruding lower lip, and a receding chin; d microdontia; e micrognathia; f redundant periumbilical skin
Detailed clinical features of the proband and the other affected individuals are summarized in Table 1.
WES variant selection and segregation analysis of the candidate variants identified
Variants obtained by WES analysis of the proband were prioritized based upon a dbSNP frequency <0.5, nucleotide and amino acid conservation, and by in silico analyses using online databases. No variants were present in the known ARS genes PITX2 and FOXC1. Therefore, candidate variants present in known genes involved in autosomal dominant anterior segment defects and glaucoma [1] (Table 2) were screened for segregation in the family.
A novel, heterozygous missense variant (c.877A>G; p.Lys293Glu) in the PRDM5 gene was found to segregate with the disease in the family (Fig. 1). The variant (c.877A>G; p.Lys293Glu) was absent in population matched controls and is also absent in the Exome Variant Server database (http://eversusgs.washington.edu/EVS/) and in the 1000 Genomes Project (http://browser.1000genomes.org/index.html). The variant was predicted to be pathogenic by most in silico analyses and is present in a Cys2His2 zinc finger protein domain of PRDM5. The lysine residue affected by the variant (c.877A>G; p.Lys293Glu) is conserved in all species analyzed (Fig. 3).
Protein structure prediction
Hope project analysis predicted that the lysine residue at position Lys293 is located in the linker between two Zn-finger domains (Fig. 4). The side chain seems to be completely exposed to the exterior surface. The lysine residue is large in size, is basic, and is positively charged, while the mutant glutamic acid (Glu) is smaller in size, is acidic, and is negatively charged. The wild-type Lys residue might have been involved in electrostatic interactions with other domains of the same protein, or with other molecules in a complex. The mutation introduces a residue with a sidechain that is smaller and that carries an opposite charge, and therefore, the interactions may be lost.
Discussion
In the current study, WES identified a novel pathogenic missense variant (c.877A>G; p.Lys293Glu) in the PRDM5 gene segregating with the disease in a family with autosomal dominant ARS. Previously, variants in the PRDM5 gene have been identified in autosomal recessive brittle cornea syndrome (BCS), and an enrichment of potentially pathogenic heterozygous variants has been observed in keratoconus [4, 14]. In BCS patients carrying PRDM5 mutations, a relationship was noted between the zygosity of the mutation (homozygous/heterozygous) and the clinical features of the patients and age of onset of keratoconus. Heterozygous carriers of PRDM5 mutations have mildly reduced central corneal thickness, mild keratoconus, and blue sclera compared to individuals with homozygous mutations, who have more severe features. Individuals with BCS who carry a homozygous deletion of exons 9–14 have an earlier age of onset and more severe keratoconus compared to individuals carrying a mutation heterozygously [4]. A recent study proposed that heterozygous variants in PRDM5 and ZNF469 predisposed the patients toward the development of isolated keratoconus [15]. These studies support that a range of ocular phenotypes are associated with variants in the PRDM5 gene, and the current study extends the spectrum to autosomal dominant ARS.
PRDM5 localizes to human chromosome 4q26 and encodes a member of the family of PRDM proteins [16]. PRDM proteins constitute a family of transcription regulators characterized by the presence of variable number of zinc finger repeats typically involved in protein–DNA or protein–protein interaction [17], and an N-terminal PR domain which shares similarity to the SET domain of histone methyltransferases. PRDM proteins play a vital role in gene expression regulation, either directly or indirectly by modifying the structure of chromatin, i.e., through the intrinsic methyltransferase activity, or via the recruitment of chromatin remodeling complexes, respectively. PRDM proteins typically display tissue-specific patterns of expression and are often involved in the differentiation of specific cell lineages [18].
PRDM5 is involved in the development and maintenance of the extracellular matrix (ECM), which explains its involvement in BCS and ARS syndromes, which represent multisystemic connective tissue disorders. Quantitative PCR analysis of fibroblast RNA from BCS patients and control individuals identified significant differences in genes involved in the ECM. In particular, genes encoding fibrillar collagens (e.g., COL4A1 and COL11A1), connective tissue components (e.g., HAPLN1), and molecules regulating cell migration and adhesion (e.g., EDIL3 and TGFB2) were significantly downregulated. EDIL3, HAPLN1, and COL11A1 each demonstrated a greater than 30-fold decrease in mutant lines relative to controls [4].
A phenotypic overlap is also present to some extent in BCS and ARS. For example, in BCS, sensorineural hearing loss is a prominent feature and was observed in families with mutations in PRDM5 described previously by Burkitt Wright et al. 2011 [4]. In the current study, patients V:1 and V:2 with ARS, a progressive sensorineural hearing loss was apparent at young age and became severe in adulthood, especially in individual V:2. In addition to this, most of the affected individuals with BCS described previously had experienced hip problems, similar to the patients V:1 and V:2 with ARS in current study. The central corneal thickness is reduced in patients with BCS, which is also reported previously in patients with ARS who underwent a corneal transplantation and carry mutations in PITX2 gene [19].
In zebrafish, Prdm5 expression was observed in specific tissues including intestinal mucosa, ventral spinal cord and ciliary zone by in situ hybridization [16]. In addition, Prdm5 is highly expressed in the osteoblast region of developing bones in mice [20]. Loss of Prdm5 results in delayed ossification, involving a prominent impairment in the assembly of fibrillar collagens. The presence of Prdm5 is therefore vital for the proper assembly of the osteoblastic extracellular matrix [20].
To study the role of Prdm5 during zebrafish development, both loss of function and gain of function approaches have been used. Two morpholino oligonucleotides were designed to obtain a depletion of Prdm5 protein: one targeting the region comprising the start codon (ATGmo), and one targeting the exon1/intron1 splice site (splice blocking or SBmo). The ATGmo induced cyclopia, while the SBmo induced closer, smaller eyes and marked axial mesendodermal defects in the jaw, heart, and blood [16]. This is in line with the eye, jaw, and heart defects and hip anomalies seen in individuals affected by ARS.
Other genes involved in ARS, FOXC1 [1, 6, 7], and PITX2 [5] also encode transcription factors. In addition, mutations in transcription factor PITX3 have been identified in anterior segment dysgenesis and congenital cataracts [5, 21]. These studies, together with the results presented here, support an important role for transcription factors and regulators in the development of anterior segment defects.
Notably, nearly all homozygous PRDM5 mutations detected in BCS patients disrupt the open reading frame, suggesting that BCS is associated with loss of function of PRDM5 [4]. In this study, a heterozygous missense variant in the linker region between two zinc finger domains of PRDM5 was identified in autosomal dominant ARS. Although the exact mechanism of action of this variant is unknown, it might have a dominant negative effect, leading to distinct ocular and extraocular features than observed in BCS. Mutations in the PITX2 gene are associated with autosomal dominant ARS, but both autosomal dominant and recessive mutations have been identified in another anterior segment defect (i.e., ring dermoid of the cornea) [22]. Both PRDM5 and PITX2 are involved in Wnt signaling pathways and are involved in the development of multiple organs [16, 23]. This suggests that PRDM5 and PITX2 mutations can lead to a spectrum of syndromic and nonsyndromic anterior segment defects, with autosomal dominant and recessive inheritance patterns. In summary, we describe a novel association of a PRDM5 missense variant with autosomal dominant ARS.
References
Ito YA, Walter MA (2014) Genomics and anterior segment dysgenesis: a review. Clin Experiment Ophthalmol 42:13–24
Tumer Z, Bach-Holm D (2009) Axenfeld-Rieger syndrome and spectrum of PITX2 and FOXC1 mutations. Eur J Hum Genet 17:1527–1539
Idrees F, Vaideanu D, Fraser SG, Sowden JC, Khaw PT (2006) A review of anterior segment dysgeneses. Surv Ophthalmol 51:213–231
Burkitt Wright EM, Spencer HL, Daly SB, Manson FD, Zeef LA, Urquhart J, Zoppi N, Bonshek R, Tosounidis I, Mohan M, Madden C, Dodds A, Chandler KE, Banka S, Au L, Clayton-Smith J, Khan N, Biesecker LG, Wilson M, Rohrbach M, Colombi M, Giunta C, Black GC (2011) Mutations in PRDM5 in brittle cornea syndrome identify a pathway regulating extracellular matrix development and maintenance. Am J Hum Genet 88:767–777
Semina EV, Reiter R, Leysens NJ, Alward WL, Small KW, Datson NA, Siegel-Bartelt J, Bierke-Nelson D, Bitoun P, Zabel BU, Carey JC, Murray JC (1996) Cloning and characterization of a novel bicoid-related homeobox transcription factor gene, RIEG, involved in Rieger syndrome. Nat Genet 14:392–399
Mears AJ, Jordan T, Mirzayans F, Dubois S, Kume T, Parlee M, Ritch R, Koop B, Kuo WL, Collins C, Marshall J, Gould DB, Pearce W, Carlsson P, Enerback S, Morissette J, Bhattacharya S, Hogan B, Raymond V, Walter MA (1998) Mutations of the forkhead/winged-helix gene, FKHL7, in patients with Axenfeld-Rieger anomaly. Am J Hum Genet 63:1316–1328
Nishimura DY, Swiderski RE, Alward WL, Searby CC, Patil SR, Bennet SR, Kanis AB, Gastier JM, Stone EM, Sheffield VC (1998) The forkhead transcription factor gene FKHL7 is responsible for glaucoma phenotypes which map to 6p25. Nat Genet 19:140–147
Stathacopoulos RA, Bateman JB, Sparkes RS, Hepler RS (1987) The Rieger syndrome and a chromosome 13 deletion. J Pediatr Ophthalmol Strabismus 24:198–203
Phillips JC, del Bono EA, Haines JL, Pralea AM, Cohen JS, Greff LJ, Wiggs JL (1996) A second locus for Rieger syndrome maps to chromosome 13q14. Am J Hum Genet 59:613–619
Werner W, Kraft S, Callen DF, Bartsch O, Hinkel GK (1997) A small deletion of 16q23.1-->16q24.2 [del(16)(q23.1q24.2).ish del(16)(q23.1q24.2)(D16S395+, D16S348-, P5432+)] in a boy with iris coloboma and minor anomalies. Am J Med Genet 70:371–376.
Riise R, Storhaug K, Brondum-Nielsen K (2001) Rieger syndrome is associated with PAX6 deletion. Acta Ophthalmol Scand 79:201–203
Sambrook J, Russell D (2000) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory, New York
Venselaar H, Te Beek TA, Kuipers RK, Hekkelman ML, Vriend G (2010) Protein structure analysis of mutations causing inheritable diseases. An e-Science approach with life scientist friendly interfaces. BMC Bioinformatics 11:548
Aldahmesh MA, Mohamed JY, Alkuraya FS (2012) A novel mutation in PRDM5 in brittle cornea syndrome. Clin Genet 81:198–199
Lechner J, Porter LF, Rice A, Vitart V, Armstrong DJ, Schorderet DF, Munier FL, Wright AF, Inglehearn CF, Black GC, Simpson DA, Manson F, Willoughby CE (2014) Enrichment of pathogenic alleles in the brittle cornea gene, ZNF469, in keratoconus. Hum Mol Genet 23:5527–5535
Meani N, Pezzimenti F, Deflorian G, Mione M, Alcalay M (2009) The tumor suppressor PRDM5 regulates Wnt signaling at early stages of zebrafish development. PLoS One 4, e4273
Fog CK, Galli GG, Lund AH (2012) PRDM proteins: important players in differentiation and disease. Bioessays 34:50–60
Kinameri E, Inoue T, Aruga J, Imayoshi I, Kageyama R, Shimogori T, Moore AW (2008) Prdm proto-oncogene transcription factor family expression and interaction with the Notch-Hes pathway in mouse neurogenesis. PLoS One 3, e3859
Kelberman D, Islam L, Holder SE, Jacques TS, Calvas P, Hennekam RC, Nischal KK, Sowden JC (2011) Digenic inheritance of mutations in FOXC1 and PITX2: correlating transcription factor function and Axenfeld-Rieger disease severity. Hum Mutat 32:1144–1152
Galli GG, Honnens de Lichtenberg K, Carrara M, Hans W, Wuelling M, Mentz B, Multhaupt HA, Fog CK, Jensen KT, Rappsilber J, Vortkamp A, Coulton L, Fuchs H, Gailus-Durner V, Hrabe de Angelis M, Calogero RA, Couchman JR, Lund AH (2012) Prdm5 regulates collagen gene transcription by association with RNA polymerase II in developing bone. PLoS Genet 8, e1002711
Aldahmesh MA, Khan AO, Mohamed J, Alkuraya FS (2011) Novel recessive BFSP2 and PITX3 mutations: insights into mutational mechanisms from consanguineous populations. Genet Med 13:978–981
Xia K, Wu L, Liu X, Xi X, Liang D, Zheng D, Cai F, Pan Q, Long Z, Dai H, Hu Z, Tang B, Zhang Z, Xia J (2004) Mutation in PITX2 is associated with ring dermoid of the cornea. J Med Genet 41, e129
Gage PJ, Suh H, Camper SA (1999) Dosage requirement of Pitx2 for development of multiple organs. Development 126:4643–4651
Acknowledgments
This study was supported by the Stichting Blindenhulp, a Shaffer grant from the Glaucoma Research Foundation, the Glaucoomfonds, Oogfonds, and the Algemene Nederlandse Vereniging ter Voorkoming van Blindheid (awarded to A. den Hollander and S. Micheal).
Conflict of interest
The authors declare no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Micheal, S., Siddiqui, S.N., Zafar, S.N. et al. Whole exome sequencing identifies a heterozygous missense variant in the PRDM5 gene in a family with Axenfeld–Rieger syndrome. Neurogenetics 17, 17–23 (2016). https://doi.org/10.1007/s10048-015-0462-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10048-015-0462-0