Abstract
Regulatory elements are more evolutionarily conserved and provide a larger mutational target than coding regions of the human genome, suggesting that mutations in non-coding regions contribute significantly to development and disease. Using a computational approach to predict gene regulatory enhancers, we found that many known and predicted embryonic enhancers cluster in genomic loci harboring development-associated genes. One of the densest clusters of predicted enhancers in the human genome is near the genes GMDS and FOXC1. GMDS encodes a short-chain mannose dehydrogenase enzyme involved in the regulation of hindbrain neural migration, and FOXC1 encodes a developmental transcription factor required for brain, heart, and eye development. We experimentally validate four novel enhancers in this locus and demonstrate that these enhancers show consistent activity during embryonic development in domains that overlap with the expression of FOXC1 and GMDS. These four enhancers contain binding motifs for several transcription factors, including the ZIC family of transcription factors. Removal of the ZIC binding sites significantly alters enhancer activity in three of these enhancers, reducing expression in the eye, hindbrain, and limb, suggesting a mechanism whereby ZIC family members may transcriptionally regulate FOXC1 and/or GMDS expression. Our findings uncover novel enhancer regions that may control transcription in a topological domain important for embryonic development.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Enhancers are gene regulatory elements—DNA regions that bind transcription factor proteins to control the timing, amplitude, and tissue specificity of gene expression [1]. The transcription factor binding sites within enhancers often have highly specific motifs, and small mutations in these binding sites can reduce or destroy binding affinity between the enhancer and the transcription factor, leading to alterations in gene expression. In a canonical example of a mutation in an enhancer causing a developmental disorder, a single point mutation in a limb-specific enhancer of the Shh (Sonic Hedgehog) gene is sufficient to cause preaxial polydactyly [2].
In this study, we demonstrate a computational approach to predict enhancers, named EnhancerFinder [3], which we used to identify a dense cluster of novel enhancers located in a topologically associating domain (TAD) containing the genes FOXC1 and GMDS. FOXC1 is a forkhead box transcription factor that is expressed in the mesenchyme of the developing somites, heart, bone, brain, and other tissues. The mouse homolog Foxc1 has been studied extensively, with Foxc1-null mutants being pre- or perinatal lethal. In humans, complete lack of FOXC1 is also typically pre- or perinatal lethal, and deletions and point mutations in FOXC1 contribute to eye and brain developmental disorders [4]. GMDS is a short-chain mannose dehydrogenase enzyme that catalyzes the production of GDP-fucose from GDP-mannose. Fucosylated glycans are known to regulate cell-cell adhesion, and thus GMDS may play a role in the regulation of cell migration and axonal pathfinding. Recent studies of the zebrafish GMDS mutants slytherin and towhead have highlighted the role of fucosylation in neural development. Slytherin mutants, which have a point mutation in GMDS, have severe hindbrain malformation and motor dysfunction that result from improper migration of hindbrain neural precursors [5]. The towhead mutation in zebrafish is less severe, resulting from a mutation of a conserved Trp residue to an Arg, manifesting in the malformation of the vagal nerve nuclei [6].
We tested four of the predicted enhancers in the FOXC1/GMDS domain and found that they consistently drive expression in developing embryonic tissues where these genes are also expressed. These enhancers contain computationally predicted binding sites for many transcription factors, including ZIC transcription factors (zinc finger of the cerebellum). We showed that removing ZIC binding sites from these enhancers significantly reduces enhancer expression in the hindbrain, eye, and limb, suggesting that ZIC genes may be involved in the transcriptional regulation of FOXC1 and/or GMDS.
Materials and methods
Enhancer identification
Enhancers tested in this study were computationally predicted using EnhancerFinder [3], a machine-learning approach that integrates thousands of genetic and epigenetic data sources to predict developmental human enhancers. The machine-learning algorithm in EnhancerFinder is unique compared to other enhancer prediction approaches, as it was trained using 700 in vivo-validated developmental enhancers from the VISTA Enhancer Browser [7]. The algorithm uses sequence motifs and patterns of transcription factor binding and histone modifications to distinguish these validated enhancers from random genomic regions or from sequences that were tested by VISTA but showed no enhancer activity. The resulting 84,301 novel enhancer candidates are predicted to be active in a variety of embryonic tissues.
Computational analysis of enhancer clusters
For every gene in the human genome (based on an October 2012 download of the UCSC Genome Browser RefGene track), we counted the number of predicted enhancers within 500 kilobases (kb) upstream or downstream of the transcription start site. While it is hard to know which gene an enhancer is actually regulating, we assumed that 1 megabase (Mb) would be a wide enough distance to capture many of the functional gene-enhancer pairs. We ranked genes based on the number of associated enhancers and thereby identified the loci with the densest clusters of enhancer candidates in the human genome.
The GREAT tool [8] was used to annotate the biological functions of the 1321 genes with the densest clusters of predicted enhancers (top 5 % of all human genes). To test if these genes are enriched for particular functions, we used the hypergeometric test, with the default GREAT settings and the “basal plus extension” association rule (proximal 5 kb upstream, 1 kb downstream, plus distal up to 100 kb).
Selection of enhancers for functional studies
We focused on a dense cluster of enhancers, which is located in the chromosome band 6p25.3 at the genomic coordinates listed below (hg19) and contains 72 enhancers within 1 Mb. Three of the enhancers (CE1–3) are located in the ∼10-kb intergenic region that falls between the neurodevelopmental genes FOXC1 and GMDS. CE4 is in the second intron of GMDS, approximately 90 kb from FOXC1. CE1–4 range in length from 995 to 1431 bp. DNA for these regions was synthesized rather than amplified via PCR, but the primers shown below have been used in a related study to amplify the same regions [3] (CE1, located at hg19 chr6:1614904-1616335, forward primer AGACCCCTGTTAGTTTCGCT and reverse primer ATTAGCTGATTCCCCGCCAT; CE2, located at chr6:1616341-1617336, forward primer AAATAGCCTCTGTAAAAAGCTTTAGG and reverse primer GACTGACACAGTCTCTTGGTCCT; CE3, located at chr6:1619847-1620844, forward primer GAGTCGAGTCCTCGGAGC and reverse primer TATGACTACGACGGCAGAGG; CE4, located at chr6:1702408-1703763, forward primer CAGTAGCTGGACTCCGACTC and reverse primer ACTTCCACCCAGCACAGAAA). Wild-type enhancer sequences consisted of the human reference genome (hg19) for each region indicated. ZIC-enhancer sequences were comprised of the reference genome sequence with all computationally predicted ZIC binding sties removed. Position weight matrices for ZIC1 and ZIC3 from the April 2011 release of the TRANSFAC database of experimentally derived transcription factor binding motifs were used to identify ZIC binding sites. The consensus sequence of ZIC binding sites is GGGGTGGTC.
Associating enhancers with genes
To link enhancers to potential target genes, we used published TADs in human embryonic stem cells. TADs are stable features of mammalian genomes that delineate regions of local 3D chromatin interactions, and the boundaries of these domains remain largely consistent across cell types and between species [9]. The topological domain that contains FOXC1, GMDS, and the predicted enhancers is defined by chromosomal position chr6:1400001-2680000 (6p25.3-6p25.2) (hg19) and spans 1.28 Mb.
Transgenic mouse enhancer assay
Mouse enhancer assays were carried out in transient transgenic mouse embryos generated by pronuclear injections of enhancer constructs into FVB embryos. Human DNA sequences were inserted upstream of the Hsp68 minimal promoter and a lacZ reporter gene. The embryos were collected and X-gal stained at E11.5. In this assay, the minimal promoter should only activate the lacZ reporter gene when the candidate enhancer region effectively recruits and binds transcriptional machinery. Following standard annotation procedures [7], we required that consistent expression patterns be present in three or more embryos to consider the candidate region an enhancer. This assay includes two negative controls: the empty vector injection and PCR-negative embryos of injected constructs. Background expression of these controls is considered in our annotation criteria. Neither of the controls exhibits the expression patterns seen in the CEs. CE1, 2, 3, and 4 all met this annotation criterion. Transgenic mice were generated by Cyagen Biosciences, whose facility meets and often exceeds animal health and welfare guidelines. Animals were euthanized using techniques recommended by the American Veterinary Medical Association. All procedures were carried out in line with Gladstone Institutes and University of California guidelines.
Identification of upstream regulators of FOXC1/GMDS
Genome-wide locations of predicted transcription factor binding sites were generated using the Find Individual Motif Occurrences (FIMO) tool from the MEME suite of bioinformatics tools [10], based on the April 2011 release TRANSFAC database of experimentally derived transcription factor binding motifs [11], with a FIMO score threshold of 10e−5. We intersected these predicted binding sites with the genomic coordinates of the four enhancers using the IntersectBed tool from the BedTools suite of bioinformatics tools [12].
To determine which genes were expressed in the developing brain, we used the fetal brain data from the GNF Atlas2 database [13]. The fetal brain data included in GNF Atlas2 is based on a Clontech pooled sample of normal whole brains from 59 spontaneously aborted male and female Caucasian fetuses, ages 20–33 weeks. All array data was mapped to RefSeq gene names (RefGene track downloaded from the UCSC Genome Browser January 2014). Genes with an expression score greater than or equal to 64 were considered expressed. To assess known gene expression patterns in the developing mouse, we viewed images of in situ hybridizations of E11.5 and E14.5 mouse embryos from the Allen Brain Map (http://developingmouse.brain-map.org/) and the Eurexpress Transcriptome Atlas (http://www.eurexpress.org/ee/). To assess known gene expression in the developing human brain, we viewed RNA-seq data from the BrainSpan Atlas of the Developing Human Brain (http://www.brainspan.org/). Genes with RPKM (reads per kilobase per million) values greater than 1 were considered to be expressed [14].
Results
EnhancerFinder is a computational tool that predicts developmental enhancers based on positive examples of biologically active developmental enhancers [7] and negative examples from genomic background. This method uses a multiple kernel learner (similar to a support vector machine) and characterizes genomic regions through an integrated profile of a large number of genetic and epigenetic data sources. Using in vivo-validated examples to train EnhancerFinder and integrating hundreds of sequence motifs and functional genomics experiments make this approach more accurate at identifying biologically active enhancers compared to other approaches [3]. For this study, we started with the 84,301 candidate developmental enhancers predicted by EnhancerFinder across the human genome. We then examined the genome-wide distribution of EnhancerFinder’s predicted enhancers and found that they cluster near loci that contain important developmental genes. Since developmentally active genes typically rely on tight regulation to exhibit robust spatio-temporal expression patterns, these enhancers likely play a role in coordinating normal development. Genes with the highest number of nearby enhancers in the human genome (Supplemental Table 1) are enriched for several biological functions related to development including epithelial cell development, arterial development, and dorsal/ventral neural tube patterning.
Many genes essential for normal development fall within clusters of EnhancerFinder’s predicted developmental enhancers. Neighboring genes FOXC1 and GMDS are found in one of the densest enhancer clusters in the genome, with 72 predicted nearby enhancers over a 1-Mb range. These genes and 104 predicted enhancers fall within a single TAD of local chromatin interactions in human embryonic stem cells [9], indicating that these regions have 3D structural interactions during embryonic development. GMDS and FOXC1 are the only two protein-coding genes fully contained within this TAD. Three long non-coding RNA genes are also encoded in this domain, as well as the 3′ end of myosin light-chain kinase MYLK4. Boundaries of topological domains are often consistent across cell types and evolution [9], suggesting that the topological domain that contains FOXC1 and GMDS is present in many developing tissues during development (Fig. 1).
The genomic landscape surrounding FOXC1 and GMDS includes four candidate enhancers tested in this study, shown in a screenshot from the UCSC Genome Browser. The top panel shows the entire topological domain that includes FOXC1 and GMDS, highlighted in green. The smaller region surrounding the four candidate enhancers is highlighted in blue and shown in greater detail in the lower panel. The genomic location, genes, and candidate enhancers are shown along with publicly available data tracks of conservation (100 Vertebrates Basewise Conservation by PhyloP) and ENCODE data of enhancer-related histone modifications H3K27ac and H3K4me1, and transcriptional activation-related histone modification H3K4me3 (“layered” view of seven cell lines including GM12878, H1-hESC, HSMM, HUVEC, K562, NHEK, and NHLF)
We tested seven candidate enhancer (CE) regions and validated five novel developmental enhancers near FOXC1 and GMDS using a transgenic mouse enhancer assay. We saw that these five enhancers are active at E11.5 in various embryonic tissues. We chose embryonic day E11.5 as it is an active stage of brain patterning and development and is 1 day prior to when differentiated structures are apparent [15]. Figure 2 shows representative images of CE1–4 from the transgenic mouse assay, as whole embryos, highlighting enhancer activity in different tissues. Associated tables detail the expression patterns of these CEs in the transgenic enhancer assay. A given anatomical region of the embryos was noted in bold when it showed expression in greater than half of the X-gal-positive (+) embryos. For instance, CE1 showed expression in the developing limb in three out of five X-gal-positive embryos (Fig. 2). Additional tissues that showed expression in more than half of the X-gal-positive embryos included the eye (CE1, 2), the spinal cord (CE1–4), and the midbrain (CE1). Expression in the hindbrain neural tube was also seen in embryos for each enhancer, although it was seen most frequently in CE4 (Figs. 2a, 3a). Interestingly, a few embryos had expression in the cortical mesenchyme (CE1, 3, 4) where Foxc1 is expressed endogenously (see Figs. 2b, 3b), although this expression was not consistent across many embryos. CE4 is more likely to be regulating GMDS as GMDS is expressed in the developing hindbrain neural tube, whereas Foxc1 is restricted to the surrounding mesenchyme (Fig. 2c, d). CE5 showed consistent expression in the eye (data not shown).
a An analysis of four computationally identified enhancers, with annotated tissue expression in the E11.5 mouse embryo. Tissues are noted in bold if they have lacZ expression half or more of the embryos for a given enhancer. b An E11.5 foxc1-lacZ embryo, X-gal stained. Arrowhead notes cortical mesenchymal expression. E14.5 in situ hybridizations of c foxc1 and d gmds in the E14.5 mouse brain. Arrowhead notes mesenchymal expression in c and hindbrain neural expression in d. Data from the Eurexpress transcriptome atlas (http://www.eurexpress.org/ee/)
The four identified CEs contain binding motifs for hundreds of transcription factors. To better understand the region’s transcriptional regulation in brain development, we filtered this list to include transcription factors and co-factors expressed in the developing brain that have at least 12 predicted binding motifs in at least one of the enhancers. We identified a large number of potential transcriptional regulators, widening the number of potential transcriptional regulators of FOXC1/GMDS expression. The neurodevelopmental transcription factors ZIC1 and ZIC3 are amongst the genes with the highest numbers of predicted binding motifs in the CEs (Table 1). Figure 4a shows the enhancer landscape near FOXC1/GMDS and the location of predicted ZIC binding sites. ZIC genes, including Zic1 and Zic4, are known to be active in the developing hindbrain [16], suggesting that they may regulate GMDS hindbrain expression.
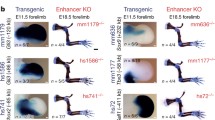
a An image from the UCSC Genome Browser showing the enhancer landscape near FOXC1 and GMDS. The top panel shows all four CEs. The lower panel is zoomed in to the regions immediately surrounding the enhancers, to highlight the ZIC1 binding sites. b E11.5 whole-mount images of ZIC mutants CE1–3. CE4 only yielded two X-gal-positive embryos and was thus not used in the analysis. Tissues that have lacZ expression in at least half of the X-gal-positive embryos in WT but not in ZIC enhancers are noted in red. Arrowheads mark regions of interest
To further investigate the role of ZIC proteins in the regulation of our identified enhancers, we made mutant enhancer constructs that lacked ZIC binding sites. We then used our transgenic mouse enhancer assay to test whether removing these sites affects expression noted in Fig. 2. We found diminished neural expression in three of the four brain CEs (Fig. 4). For the mutant CE4 construct, we did not recover enough embryos to make substantive conclusions (only two X-gal-positive embryos were recovered). Of note, many regions where at least half of the X-gal-positive embryos had expression in with the wild-type (WT) enhancer no longer had robust expression. This included expression in the eye (CE1 and 2), limb (CE1), spinal cord (CE2 and 3), and midbrain (CE1). Also, a number of regions gained expression that was not seen in the WT embryos, including the branchial arch and facial mesenchyme. While mouse ZIC1 or 3 do not appear to be expressed in these structures, ZIC2 is expressed in the developing upper and lower jaws (Figure S1). Thus, ZIC2 may be inhibiting craniofacial expression in the WT context and the lack of ZIC binding sites in the mutant enhancers may lead to disinhibition and activation of expression in craniofacial tissues.
Although the CEs are located very close to FOXC1 and GMDS, they may also regulate other nearby genes. FOXF2 and FOXQ1 are located 215 and 300 kb, respectively, upstream of FOXC1 and the CE cluster. These genes are expressed in many of the same mouse tissues as the CEs (as seen in in situ images of E11.5 mouse embryos from the Allen Brain Map). Although these other potential target genes are not in the same TAD as the CEs in human embryonic stem cells, the genomic distance separating them from the CE cluster is well within the range of enhancer function. Together, FOXC1, GMDS, FOXF2, and FOXQ1 may represent loci for enhancer-driven changes in gene expression that could affect the developing embryo.
Discussion
Here, we have applied a novel computational approach for discovering enhancers, called EnhancerFinder. We found that many enhancers cluster around developmentally relevant genes, such as transcription factors that are necessary for controlling cell identity. By identifying novel enhancers, EnhancerFinder provides new avenues of investigation regarding the regulation of gene expression during embryonic development. We characterize a cluster of predicted enhancers around the genes FOXC1 and GMDS, both of which are known to regulate diverse developmental processes in multiple tissues. We found that these enhancers are contained within a single TAD, suggesting a common underlying structural constraint that may link these enhancers to FOXC1 and GMDS. With the large number of EnhancerFinder-predicted regions that cluster within this domain, the FOXC1 and GMDS locus appears to be densely packed with regulatory elements, meriting future investigation.
Using an embryonic mouse transgenic assay, we show that a set of novel enhancers drives expression in many of the regions that express FOXC1 and/or GMDS. We also provide sequence-based evidence that ZIC protein family members such as ZIC1 are likely candidate regulators of expression from these enhancers. To support this, we show that the tissue-specific activity of these enhancers is dependent upon the presence of ZIC transcription factor binding motifs in the enhancers.
A number of developmental processes are regulated by GMDS and FOXC1. GMDS controls the production of fucosylated glycans, which in turn regulates cell migration and adhesion, as well as the formation of axonal projections and synapses [4, 5, 17]. Notch signaling is also regulated via the fucosylation of EGF-like repeats in the Notch receptor, the disruption of which was shown to contribute, at least in part, to the slytherin GMDS mutant phenotype [5]. FOXC1 controls the development of a number of tissues, including the mesenchyme that surrounds the nervous system, as well as the somatic mesenchyme, heart, kidney, and eye. FOXC1 mutations in humans result in developmental malformations of the cerebellum and eye, Axenfeld-Rieger syndrome, and Dandy-Walker malformation [4]. These disorders likely result from defective meningeal development and disrupted mesenchymal-neuroepithelial interactions during embryonic development.
The specific role of regulatory elements in the FOXC1/GMDS locus during brain development is unclear. Specifically, it is unknown what role non-coding mutations around the FOXC1/GMDS locus might play in developmental disorders such as Dandy-Walker syndrome. A small number of the transgenic embryos tested in this study showed mesenchymal enhancer expression suggestive of the FOXC1-like expression, although the number was too few to make any definitive conclusions. Neuronal expression was seen more consistently, similar to neuronal GMDS expression in the embryonic brain. Given the role of GMDS in hindbrain development in zebrafish [5, 6], it is possible that mutations in enhancers surrounding the FOXC1/GMDS locus may disrupt GMDS expression and thus cerebellar development. Also, ZIC genes in humans and in mice have been shown to be involved in Dandy-Walker cerebellar malformations [16, 18], which is interesting given the particularly high number of ZIC binding motifs in enhancers near FOXC1 and GMDS. Thus, these novel enhancers provide mechanistic insight into means by which these two genes are regulated in a variety of tissues in the developing embryo. The initial characterization of the FOXC1/GMDS regulatory environment we present here may help inform further areas of research regarding developmental malformations that result from mutations in this locus.
References
Noonan JP, McCallion AS (2010) Genomics of long-range regulatory elements. Annu Rev Genomics Hum Genet 11:1–23. doi:10.1146/annurev-genom-082509-141651
Maas SA, Fallon JF (2005) Single base pair change in the long-range Sonic hedgehog limb-specific enhancer is a genetic basis for preaxial polydactyly. Dev Dyn 232(2):345–348. doi:10.1002/dvdy.20254
Erwin GD, Oksenberg N, Truty RM, Kostka D, Murphy KK, Pollard KS, Ahituv N, Capra JA (2014) Integrating diverse datasets improves developmental enhancer prediction. PLoS Comput Biol 10(6):e1003677. doi:10.1371/journal.pcbi.1003677
Aldinger KA, Lehmann OJ, Hudgins L, Chizhikov VV, Bassuk AG, Ades LC, Krantz ID, Dobyns WB, Millen KJ (2009) FOXC1 is required for normal cerebellar development and is a major contributor to chromosome 6p25.3 Dandy-Walker malformation. Nat Genet 41(9):1037–1042. doi:10.1038/ng.422
Song Y, Willer JR, Scherer PC, Panzer JA, Kugath A, Skordalakes E, Gregg RG, Willer GB, Balice-Gordon RJ (2010) Neural and synaptic defects in slytherin, a zebrafish model for human congenital disorders of glycosylation. PLOS One 5(10):e13743. doi:10.1371/journal.pone.0013743
Ohata S, Kinoshita S, Aoki R, Tanaka H, Wada H, Tsuruoka-Kinoshita S, Tsuboi T, Watabe S, Okamoto H (2009) Neuroepithelial cells require fucosylated glycans to guide the migration of vagal motor progenitors in the developing zebrafish hindbrain. Development 136(10):1653–63. doi:10.1242/dev.033290
Visel A, Minovitsky S, Dubchak I, Pennacchio LA (2007) VISTA enhancer browser—a database of tissue-specific human enhancers. Nucleic Acids Res 35(Database issue):D88–D92. doi:10.1093/nar/gkl822
McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G (2010) GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 28(5):495–501. doi:10.1038/nbt.1630
Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B (2012) Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485:376–380. doi:10.1038/nature11082
Grant CE, Bailey TL, Noble WS (2011) FIMO: scanning for occurrences of a given motif. Bioinformatics 27(7):1017–1018. doi:10.1093/bioinformatics/btr064
Wingender E, Chen X, Hehl R, Karas H, Liebich I, Matys V, Meinhardt T, Pruss M, Reuter I, Schacherer F (2000) TRANSFAC: an integrated system for gene expression regulation. Nucleic Acids Res 28(1):316–319
Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26(6):841–842. doi:10.1093/bioinformatics/btq033
Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, Cooke MP, Walker JR, Hogenesch JB (2004) A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A 101(16):6062–6067. doi:10.1073/pnas.0400782101
BrainSpan (2013) Transcriptome profiling by RNA sequencing and exon microarray. Technical White Paper
Rossant J, Tam PPL (2002) Mouse development: patterning, morphogenesis, and organogenesis. Academic, San Diego
Blank MC, Grinberg I, Aryee E, Laliberte C, Chizhikov VV, Henkelman RM, Millen KJ (2011) Multiple developmental programs are altered by loss of Zic1 and Zic4 to cause Dandy-Walker malformation cerebellar pathogenesis. Development 138(6):1207–1216. doi:10.1242/dev.054114
Panzer, JA, Gibbs SM, Dosch R, Wagner D, Mullins MC, Granato M, Balice-Gordon RJ (2005) Neuromuscular synaptogenesis in wild-type and mutant zebrafish. Develop Biol 285(2):340–57. doi:10.1016/j.ydbio.2005.06.027
Grinberg I, Northrup H, Ardinger H, Prasad C, Dobyns WB, Millen KJ (2004) Heterozygous deletion of the linked genes ZIC1 and ZIC4 is involved in Dandy-Walker malformation. Nat Genet 36:153–1055
Acknowledgments
We would like to thank Nir Oksenberg for the help in imaging mouse embryos, Caroline Miller for histology services with mouse brain sections, Sean Thomas for sharing predictions of genome-wide locations of transcription factor binding sites, Julie Siegenthaler for providing a foxc1-lacZ embryo, and Kate Lovero for helpful discussion. Funding was provided by grants from the National Heart, Lung, and Blood Institute (NHLBI) (#HL098179), a University of California Achievement Awards for College Scientists (ARCS) Scholarship, a T32 training grant to the UCSF Pediatric Endocrinology Department (#5T32DK007161-41), a gift from the San Simeon Fund, and institutional funds from the J. David Gladstone Institutes. Transgenic mice were generated by Cyagen Biosciences, Inc.
Author information
Authors and Affiliations
Corresponding author
Additional information
Genevieve D. E. Haliburton and Gabriel L. McKinsey contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Figure 1
An E14.5 in situ hybridization of Zic2, from the Eurexpress Transcriptome Atlas. Arrow heads note expression in the developing upper and lower jaws (http://www.eurexpress.org/ee/). (PDF 10390 kb)
Table S1
Genes with the top 5 % highest number of enhancers within a 1-Mb window (500 kb upstream and downstream) of the transcription start site. The genes are listed in alphabetical order along with the number of nearby enhancers. The number of nearby enhancers ranges from 61 to 106. (TXT 14 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Haliburton, G.D.E., McKinsey, G.L. & Pollard, K.S. Disruptions in a cluster of computationally identified enhancers near FOXC1 and GMDS may influence brain development. Neurogenetics 17, 1–9 (2016). https://doi.org/10.1007/s10048-015-0458-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10048-015-0458-9