Abstract
Purpose
Inguinal hernia repair is one of the most common procedures in general surgery. Males are seven times more likely than females to develop a hernia and have a 27 % lifetime ‘risk’ of inguinal hernia repair. Several studies have demonstrated that a positive family history is an important risk factor for the development of primary inguinal hernia, which indicates that genetic factors may play important roles in the etiology of the disease. So far, the contribution of genetic factors and underlying mechanisms for inguinal hernia remain largely unknown. The aim of this study was to investigate a multiplex Estonian family with inguinal hernia across four generations.
Methods
The whole-exome sequencing was carried out in three affected family members and subsequent mutation screening using Sanger sequencing was performed in ten family members (six affected and four unaffected).
Results
Whole-exome sequencing in three affected family members revealed a heterozygous missense mutation c.88880A>C (p.Lys29627Thr; RefSeq NM_001256850.1) in the highly conserved myosin-binding A-band of the TTN gene. Sanger sequencing demonstrated that this mutation cosegregated with the disease in this family and was not present in ethnically matched control subjects.
Conclusion
We report that missense variant in the A-band of TTN is the strongest candidate mutation for autosomal-dominant inguinal hernia with incomplete penetrance.
Similar content being viewed by others
Introduction
Although the lifetime risk for inguinal hernia operation is estimated to be 27 % for men and 3 % for women [1] resulting in vast number of patients undergoing inguinal hernia repair, our knowledge of the etiology of the disease is insufficient. It is estimated that over 700,000 groin hernias are repaired annually both in the United States and Europe; specific rates vary between countries from 100 to 300 per 100,000 population per year [2, 3].
Inguinal hernia is often classified into two subtypes based on their localizations as direct and indirect [4, 5]. Indirect inguinal hernias emerge from the inguinal ring, whereas direct inguinal hernias arise from Hesselbach’s triangle [5, 6]. Indirect hernia is more common during adolescent age and direct hernia in adults [7].
According to the European Hernia Society guidelines on the treatment of inguinal hernia, smokers, patients with patent processus vaginalis, after an open appendectomy and prostatectomy, with ascites, on peritoneal dialysis, after long-term hard work or with chronic obstructive pulmonary disease have an increased risk of inguinal hernia development [8]. Connective tissue disorders, such as Ehlers–Danlos and Marfan syndromes, and systemic collagen subtype imbalance have also been associated with an increased risk of hernia [9, 10]. Positive family history represents an important risk factor for development of primary inguinal hernia and suggests the role of genetic factors in the etiology of the disease [5, 11, 12]. There are only a few studies that have investigated genetic predisposition to inguinal hernia and therefore, the contribution of genetic factors still remains largely unknown [11, 13, 14]. So far, autosomal dominant with incomplete penetrance and multifactorial inheritance are two mainly suggested inheritance patterns [13, 15].
In this paper, we present the results of an investigation on inguinal hernia in a large multiplex Estonian family using a whole-exome sequencing strategy followed by validation with Sanger sequencing.
Materials and methods
Subjects
Ethical approvals for the study were obtained from the Ethics Review Committee on Human Research of the University of Tartu. All participants in this study were recruited from The Estonian Biobank cohort [16]. An informed consent form was signed prior to enrollment in this study by all participants. Genomic DNA was extracted by standard procedures from peripheral lymphocytes of affected individuals and unaffected siblings. All participants have filled the health questionnaires and the GPs performed a standardized health examination of the participants. Inguinal hernia patients’ diagnoses were confirmed according to medical records. The characteristics of all individuals from an Estonian family who participated in this study are in Table 1. Inguinal hernia was diagnosed through five generations. Three patients had indirect hernias (hernias that occurs lateral to the epigastric vessels) and two had direct inguinal hernias (hernias that occur medial to the epigastric vessels). An average age at the disease onset was 20.4 years (SD ± 27.88). All patients were non-smokers; calculated body mass indexes were between 20.01 and 30.07 kg/m2.
Exome sequencing
Three affected family members (IV:3, IV:6 and IV:16; Fig. 1) were analyzed by the whole-exome sequencing as described previously [17]. Briefly, exome capture was performed using the Agilent SureSelect Human All Exon 50 Mb kit (Agilent Technologies Inc., Santa Clara, CA). The captured DNA was sequenced on Illumina HiSeq 2000 according to the manufacturer’s protocol. 100-bp paired-end sequence reads were aligned to the human reference genome (hg19, GRCh37) using the Burrows–Wheeler Aligner (BWA, version 0.6.1) [18]. Single-nucleotide substitutions (SNVs) and small indel variants were called with SAMtools (version 0.1.18), Picard tools (version 1.60; http://picard.sourceforge.net/), and a Genome Analysis Toolkit (GATK, version 1.5.21) and annotated with in-house scripts [19, 20]. Retained SNVs and indels with Phred-like quality scores of >20 were filtered against dbSNP138 and the 1000 Genomes project to exclude previously identified variants, and a comparison with our in-house database of Estonian exomes and full genomes was then performed to further reduce the number of candidate mutations. We focused on heterozygous SNVs and insertions/deletions affecting coding regions or canonical splicing sites. PolyPhen-2 and MutationTaster software tools were used to predict the functional effects of mutations.
Pedigree of the analyzed family and confirmation of the TTN mutation by Sanger sequencing. Symbols marked with capital A (dark blue) indicate individuals with inguinal hernia. Symbols marked with capital U (green) indicate unaffected individuals. Symbols marked with yellow indicate individuals whose disease status in unknown. A plus sign indicates individuals whose DNA was available for this study. Individuals IV:3, IV:6 and IV:16 underwent exome sequencing; individuals III:2, IV:3, IV:6, IV:7, V:4, V:6, V:7, IV:16, IV:17, V:14 underwent Sanger sequencing
Segregation analysis
Sanger sequencing was used to confirm that mutations identified by exome sequencing segregated with the disease. Primer sequences are presented in the Supplementary Table 1. Sequencing was performed using the BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, USA) according to the manufacturer’s protocol with a 3730xl DNA Analyzer (Applied Biosystems, USA). Mutation analysis was performed on ten family members (III:2, IV:3, IV:6, IV:7, V:4, V:6, V:7, IV:16, IV:17, V:14; Fig. 1; Table 1).
Linkage analysis
Linkage analysis was performed on nine family members (III:2, IV:3, IV:6, IV:7, V:4, V:6, IV:16, IV:17, V:14; Fig. 1; Table 1). All indicated samples were genotyped using an Illumina OmniExpress BeadChip (Illumina, San Diego, CA, USA). Multipoint linkage analysis was performed using MERLIN 1.1.2 [21] under the autosomal dominant inheritance model with a mutant allele frequency of 0.0001 and incomplete penetrance.
Results
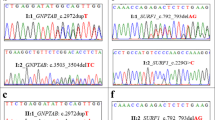
Whole-exome sequencing was performed in three affected individuals (IV:3, IV:6, IV:16; Fig. 1; Table 1); two brothers and their niece. The exomes were covered at least 50-fold and the mean read depth was 108. In each individual, between 58,194 and 61,242 single nucleotide variants (SNVs) and 4641-6460 small insertions–deletions were identified. In each individual, 613-832 SNVs and 655-998 indels were not present in dbSNP138 or the 1000 Genomes project database. After exclusion of variants found in healthy individuals, and by further filtering against in-house exomes database, three heterozygous mutations remained for segregation analysis (Fig. 2a). Sanger sequencing confirmed that only heterozygous missense mutation (c.88880A>C; RefSeq NM_001256850.1; p.Lys29627Thr) in exon 339 within Ig-like 135 domain of TTN co-segregated with inguinal hernia (Fig. 2a, b). This mutation was present in all affected individuals who were descendants of III:2 or III:4, and in an unaffected carrier female V:14 (Fig. 1). The mutation was not detected in unaffected family members V:6, IV:17, IV:7 and in affected individual V:7. This mutation was not present in Estonian control exomes of 334 individuals with unrelated conditions and in full genomes of 2340 Estonians. Both PolyPhen-2 and MutationTaster predicted this mutation as probably damaging or disease causing. The Lys29627 position is highly conserved (phyloP score 5.199) in TTN proteins from other vertebrate species (Fig. 2c).
a Validation of the identified c.88880A>C mutation. b Sanger sequencing confirmed the A to C substitution (indicated by an arrow) corresponding to c.88880A>C in exon 339 (NM_001256850.1), shown in affected individual IV:6 and unaffected individual IV:17. c Multiple sequence alignment of TTN sequences across species. Asterisks mark the positions of fully conserved amino acids. The Lys29627 position is marked with black border
Genome-wide linkage analysis revealed no significant logarithm of odd scores.
Discussion
Although inguinal hernia repair is one of the most commonly performed procedures in general surgery, the pathogenic mechanisms of inguinal hernias are not completely understood. Inguinal hernia has been proposed to be hereditary [11, 22], but the possible inheritance pattern is uncertain yet and there are no reported genetic studies for humans that could be considered as whole-genome level or genome-wide.
In the current study, whole-exome sequencing has revealed a heterozygous missense mutation c.88880A>C (p.Lys29627Thr) in the TTN gene, encoding for titin, that co-segregated with inguinal hernia in an Estonian multiplex family. This mutation was present in all affected individuals (except V:7, who descends from a different family tree) and also in a female carrier V:14 without onset of inguinal hernia during her lifespan. It is common that individuals with a family history of hernia may not experience any symptoms until an older age. This is supported by the fact that her two sons both have already a diagnosed inguinal hernia, unfortunately they were unavailable to be included to the current study. It is more likely that inheritance pattern in this family is autosomal-dominant with incomplete penetrance. This is consistent with previous studies focusing on genetic background of inguinal hernia that have proposed inguinal hernia to be a polygenic trait that may possibly occur as an autosomal-dominantly inherited disease with reduced penetrance in familial cases [13, 15]. The family used in the current study consists of patients with both direct and indirect hernias. It has been proposed that indirect hernia is more common during adolescent age and direct hernia in adults [7], this may suggest that the children and the adult inguinal hernia can have different etiologies [11]. Thus, although we have both types on hernias in the family used in the study, which either can be considered as the weakness of the current study or there are some additional genetic factors that could be common to the both types of inguinal hernias. Further studies with this family may provide new information.
TTN is an extremely large protein and is abundantly expressed in highest levels in heart and skeletal muscle [23], where it spans half of the sarcomere and runs from the Z-disc through the I-band and A-band all the way to the M-line [24]. The human titin gene encompasses ~300 kb on chromosome 2q24 and its coding sequence is comprised of 363 exons (ENSG00000155657). The c.88880A>C transversion is located in exon 339 within the myosin-binding A-band segment of titin and leads to the substitution of basic positively charged lysine to uncharged threonine. The A-band of titin, the largest part of the molecule arranged in 7- and 11-domain super-repeats [25], is very stable and highly conserved [26]. TTN gene is known to be involved in muscle contraction, sarcomerogenesis, and skeletal muscle myosin filaments assembly; TTN mutations have been reported as clinically associated variants for both human skeletal muscle and heart diseases. Several studies have described mutations in the A-band of titin to be associated with dilated cardiomyopathy or hereditary myopathy with early respiratory failure [27, 28]. The c.88880A>C is reported in dbSNP138 as an rs200766837; however, this variant is monomorphic in Estonian general population based on genotyping data of 4534 randomly selected individuals from the Estonian Biobank. Only a few TTN mutations that are associated with heart or skeletal muscle disease have also been identified in general population, although polymorphisms in the TTN gene are rather abundant in healthy individuals [29, 30]. The possible consequences of c.88880A>C may lead to disrupted interactions with titin-binding proteins (e.g. ANKRD1, ANK1, CAPN3), and/or misfolding of defective titin protein.
We therefore conclude that reported missense variant in the A-band of TTN is the strongest candidate mutation for autosomal dominant inguinal hernia with incomplete penetrance. Our study provides support with the previous studies suggesting that the inheritance pattern of familial inguinal hernia could be autosomal dominant. Further studies will be needed to clarify the genotype–phenotype correlations in the patients with rare deleterious mutations in TTN.
References
Primatesta P, Goldacre MJ (1996) Inguinal hernia repair: incidence of elective and emergency surgery, readmission and mortality. Int J Epidemiol 25:835–839
Burcharth J, Pedersen M, Bisgaard T et al (2013) Nationwide prevalence of groin hernia repair. PLoS ONE 8:e54367. doi:10.1371/journal.pone.0054367
Rutkow IM (2003) Demographic and socioeconomic aspects of hernia repair in the United States in 2003. Surg Clin North Am 83:1045–1051, v–vi. doi:10.1016/S0039-6109(03)00132-4
Jenkins JT, O’Dwyer PJ (2008) Inguinal hernias. BMJ 336:269–272. doi:10.1136/bmj.39450.428275.AD
Akbulut S, Cakabay B, Sezgin A (2010) A familial tendency for developing inguinal hernias: study of a single family. Hernia 14:431–434. doi:10.1007/s10029-009-0554-6
Van Wessem KJ, Simons MP, Plaisier PW, Lange JF (2003) The etiology of indirect inguinal hernias: congenital and/or acquired? Hernia 7:76–79. doi:10.1007/s10029-002-0108-7
Ein SH, Njere I, Ein A (2006) Six thousand three hundred sixty-one pediatric inguinal hernias: a 35-year review. J Pediatr Surg 41:980–986. doi:10.1016/j.jpedsurg.2006.01.020
Simons MP, Aufenacker T, Bay-Nielsen M et al (2009) European Hernia Society guidelines on the treatment of inguinal hernia in adult patients. Hernia 13:343–403. doi:10.1007/s10029-009-0529-7
Henriksen NA, Yadete DH, Sorensen LT et al (2011) Connective tissue alteration in abdominal wall hernia. Br J Surg 98:210–219. doi:10.1002/bjs.7339
Casanova AB, Trindade EN, Trindade MRM (2009) Collagen in the transversalis fascia of patients with indirect inguinal hernia: a case-control study. Am J Surg 198:1–5. doi:10.1016/j.amjsurg.2008.07.021
Burcharth J, Pommergaard HC, Rosenberg J (2013) The inheritance of groin hernia: a systematic review. Hernia 17:183–189. doi:10.1007/s10029-013-1060-4
Zöller B, Ji J, Sundquist J, Sundquist K (2013) Shared and nonshared familial susceptibility to surgically treated inguinal hernia, femoral hernia, incisional hernia, epigastric hernia, and umbilical hernia. J Am Coll Surg 217(289–99):e1. doi:10.1016/j.jamcollsurg.2013.04.020
Gong Y, Shao C, Sun Q et al (1994) Genetic study of indirect inguinal hernia. J Med Genet 31:187–192
Zhang Y, Han Q, Fan H et al (2014) Genetic analysis of the TBX2 gene promoter in indirect inguinal hernia. Hernia 18:513–517. doi:10.1007/s10029-013-1199-z
Jones ME, Swerdlow AJ, Griffith M, Goldacre MJ (1998) Risk of congenital inguinal hernia in siblings: a record linkage study. Paediatr Perinat Epidemiol 12:288–296
Leitsalu L, Haller T, Esko T et al (2014) Cohort Profile: Estonian Biobank of the Estonian Genome Center, University of Tartu. Int J Epidemiol. doi:10.1093/ije/dyt268
Nikopensius T, Saag M, Jagomägi T et al (2013) A missense mutation in DUSP6 is associated with class III malocclusion. J Dent Res 92:893–898. doi:10.1177/0022034513502790
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25:1754–1760. doi:10.1093/bioinformatics/btp324
Li H, Handsaker B, Wysoker A et al (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi:10.1093/bioinformatics/btp352
McKenna A, Hanna M, Banks E et al (2010) The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. doi:10.1101/gr.107524.110
Abecasis GR, Cherny SS, Cookson WO, Cardon LR (2002) Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30:97–101. doi:10.1038/ng786
Marshall AG, Hutchinson EO, Honisett J (1962) Heredity in common diseases. A retrospective survey of twins in a hospital population. Br Med J 1:1–6
Labeit S, Kolmerer B, Linke WA (1997) The giant protein titin. Emerging roles in physiology and pathophysiology. Circ Res 80:290–294
Krüger M, Linke WA (2009) Titin-based mechanical signalling in normal and failing myocardium. J Mol Cell Cardiol 46:490–498
Krüger M, Linke WA (2011) The giant protein titin: a regulatory node that integrates myocyte signaling pathways. J Biol Chem 286:9905–9912. doi:10.1074/jbc.R110.173260
Tskhovrebova L, Walker ML, Grossmann JG et al (2010) Shape and flexibility in the titin 11-domain super-repeat. J Mol Biol 397:1092–1105. doi:10.1016/j.jmb.2010.01.073
Toro C, Olivé M, Dalakas MC et al (2013) Exome sequencing identifies titin mutations causing hereditary myopathy with early respiratory failure (HMERF) in families of diverse ethnic origins. BMC Neurol 13:29. doi:10.1186/1471-2377-13-29
Herman DS, Lam L, Taylor MR et al (2012) Truncations of titin causing dilated cardiomyopathy. N Engl J Med 366:619–628. doi:10.1056/NEJMoa1110186
Linke WA, Hamdani N (2014) Gigantic business: titin properties and function through thick and thin. Circ Res 114:1052–1068. doi:10.1161/CIRCRESAHA.114.301286
Golbus JR, Puckelwartz MJ, Fahrenbach JP et al (2012) Population-based variation in cardiomyopathy genes. Circ Cardiovasc Genet 5:391–399. doi:10.1161/CIRCGENETICS.112.962928
Acknowledgments
We thank the patients and their family. EGCUT studies were covered by EstRC (IUT24-6, IUT20-60), EU H2020 grant no. 692145/ePerMed and Center of Excellence in Genomics and Translational Medicine (TK142 GenTransMed).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Statement of human and animal rights
Ethical approvals for the study were obtained from the Ethics Review Committee on Human Research of the University of Tartu, Estonia (No. 240/T-2 from September 15, 2014). The study was conducted according to the ethical standards of the Ethics Review Committee on Human Research of the University of Tartu, Estonia and with the Helsinki declaration and its amendments. The study did not involved animals.
Informed consent
An informed consent form was signed prior to enrollment in this study by all participants.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/), which permits any noncommercial use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Mihailov, E., Nikopensius, T., Reigo, A. et al. Whole-exome sequencing identifies a potential TTN mutation in a multiplex family with inguinal hernia. Hernia 21, 95–100 (2017). https://doi.org/10.1007/s10029-016-1491-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10029-016-1491-9