Abstract
Context
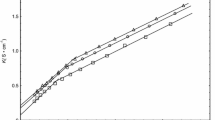
Choline-based surfactants are interesting both from the practical point of view to obtaining environmental-friendly surfactants as well as from the theoretical side since the interactions between the choline and surfactants can help to understand self-assembly phenomena in deep eutectic solvents. Although no significant change was noticed in the micelle size and shape due to the exchange of the sodium counter-ion by choline in our simulations, the adsorption of the choline cation over the micelle surface is stronger than the adsorption of the sodium, which leads to a reduction of the exposed surface area of the micelle and remarkable effects over the electrostatic potential. The choline neutralizes the surface charge of the surfactant better than sodium; however, this is partially compensated by a stronger water orientation around the SDS micelle. The balance between the contributions from the surfactant, the counter-ion, and water to the electrostatic potential leads to a complex pattern with alternate regions of positive and negative potential at the micelle/water interface which can be important to the incorporation of other charged species at the micelle surface as well as for the interaction between micelles in solution.
Methods
To evaluate the effects of the counter-ion substitution, micelles of sodium dodecyl sulfate (SDS) and choline dodecyl sulfate (ChDS) were studied and compared by means of molecular dynamics simulations in aqueous solution. In both cases, the simulations started from pre-assembled micelles with 60 dodecyl sulfate ions and 240-ns simulations were performed at NPT ensemble at T = 323.15 K and P = 1 bar using the Gromacs software with the OPLS-AA force field to describe dodecyl sulfate and choline, Åqvist parameters for sodium, and SPC model for water molecules.








Similar content being viewed by others
Data availability
Input and output files for the simulations can be provided by the author via a reasonable request by the e-mail: kalilb@ufscar.br.
References
Tanford C (1973) The hydrophobic effect: Formation of micelles and biological membranes. John Wiley & Sons, New York
Israelachvili JN (2011) Intermolecular and Surface Forces, 3rd edn. Elsevier Academic Press, San Diego
Shaban SM, Kang J, Kim D-H (2020) Surfactants: Recent advances and their applications. Compos Commun 22:100537
Holmberg K (2004) Surfactant-templated nanomaterials synthesis. J Colloid Interf Sci 274:355–364
de Moura AF, Bernardino K, Dalmaschio CJ, Leite ER, Kotov NA (2015) Thermodynamic insights into the self-assembly of capped nanoparticles using molecular dynamic simulations. Phys Chem Chem Phys 17:3820–3831
Bernardino K, de Moura AF (2013) Aggregation Thermodynamics of Sodium Octanoate Micelles Studied by Means of Molecular Dynamics Simulations. J Phys Chem B 117(24):7324–7334
Liu G, Zhang H, Liu G, Yuan S, Liu C (2016) Tetraalkylammonium interactions with dodecyl sulfate micelles: a molecular dynamics study. Phys Chem Chem Phys 18:878–885
Hantal G, Pártay LB, Varga I, Jedlovszky P, Gilányi T (2007) Counterion and Surface Density Dependence of the Adsorption Layer of Ionic Surfactants at the Vapor−Aqueous Solution Interface: A Computer Simulation Study. J Phys Chem B 111:1769–1774
Rakitin AR, Pack GR (2004) Molecular Dynamics Simulations of Ionic Interactions with Dodecyl Sulfate Micelles. J Phys Chem B 108:2712–2716
De Moura AF, Freitas LCG (2004) Molecular Dynamics simulation of the sodium octanoate micelle in aqueous solution: comparison of force field parameters and molecular topology effects on the micellar structure. Braz J Phys 34:64–72
Tang X, Koenig PH, Larson RG (2014) Molecular Dynamics Simulations of Sodium Dodecyl Sulfate Micelles in Water—The Effect of the Force Field. J Phys Chem B 118:3864–3880
Cunha RD, Ferreira LJ, Orestes E, Coutinho-Neto MD, de Almeida JM, Carvalho RM, Maciel CD, Curutchet C, Homem-de-Mello P (2022) Naphthenic Acids Aggregation: The Role of Salinity. Computation 10:170
Poghosyan AH, Arsenyan LH, Shahinyan AA (2015) Shape of Long Chain Alkyl Sulfonate Micelle upon Salt Addition: a Molecular Dynamics Study. J Surfact Deterg 18:755–760
Volkov NA, Tuzov NV, Shchekin AK (2016) Molecular dynamics study of salt influence on transport and structural properties of SDS micellar solutions. Fluid Ph Equilib 424:114–121
Sammalkorpi M, Karttunen M, Haataja M (2009) Ionic Surfactant Aggregates in Saline Solutions: Sodium Dodecyl Sulfate (SDS) in the Presence of Excess Sodium Chloride (NaCl) or Calcium Chloride (CaCl2). J Phys Chem B 113:5863–5870
Zana R (2004) Partial Phase Behavior and Micellar Properties of Tetrabutylammonium Salts of Fatty Acids: Unusual Solubility in Water and Formation of Unexpectedly Small Micelles. Langmuir 20:5666–5668
Chowdhury MR, Moshikur RM, Wakabayashi R, Tahara Y, Kamiya N, Moniruzzaman M, Goto M (2019) In vivo biocompatibility, pharmacokinetics, antitumor efficacy, and hypersensitivity evaluation of ionic liquid-mediated paclitaxel formulations. Int J Pharm 565:219–226
Zeisel SH, da Costa KA (2009) Choline: an essential nutrient for public health. Nutr Rev 67:615–623
Wei Y, Wang H, Liua G, Wang Z, Yuan S (2016) A molecular dynamics study on two promising green surfactant micelles of choline dodecyl sulfate and laurate. RSC Adv 6:84090–84097
Bernardino K, de Moura AF (2015) Surface Electrostatic Potential and Water Orientation in the presence of Sodium Octanoate Dilute Monolayers Studied by Means of Molecular Dynamics Simulations. Langmuir 31:10995–11004
Bernardino K, de Moura AF (2019) Electrostatic potential and counterion partition between flat and spherical interfaces. J Chem Phys 150:074704
Tamucci JD, Alder NN, May ER (2023) Peptide Power: Mechanistic Insights into the Effect of Mitochondria-Targeted Tetrapeptides on Membrane Electrostatics from Molecular Simulations. Mol Pharmaceutics 20:6114–6129
Gurtovenko AA, Vattulainen I (2008) Membrane Potential and Electrostatics of Phospholipid Bilayers with Asymmetric Transmembrane Distribution of Anionic Lipids. J Phys Chem B 112:4629–4634
Gurtovenko AA, Vattulainen I (2009) Calculation of the electrostatic potential of lipid bilayers from molecular dynamics simulations: Methodological issues. J Chem Phys 130:215107
Smith EL, Abbott AP, Ryder KS (2014) Deep Eutectic Solvents (DESs) and Their Applications. Chem Rev 114:11060–11082
Coutinho JA, Pinho SP (2017) Special issue on deep eutectic solvents: a foreword. Fluid Ph Equilib 448:1
Abbott AP, Capper G, Davies DL, Rasheed RK, Tambyrajah V (2003) Novel solvent properties of choline chloride/urea mixtures. Chem Commun 1:70–71
Shekaari H, Zafarani-Moattar MT, Shayanfar A, Mokhtarpour M (2018) Effect of choline chloride/ethylene glycol or glycerol as deep eutectic solvents on the solubility and thermodynamic properties of acetaminophen. J Mol Liquids 249:1222–1235
Sanchez-Fernandez A, Edler KJ, Arnold T, Heenan RK, Porcar L, Terrill NJ, Terry AE, Jackson AJ (2016) Micelle structure in a deep eutectic solvent: a small-angle scattering study. Phys Chem Chem Phys 18:14063–14073
Arnold T, Jackson AJ, Sanchez-Fernandez A, Magnone D, Terry AE, Edler KJ (2015) Surfactant Behavior of Sodium Dodecylsulfate in Deep Eutectic Solvent Choline Chloride/Urea. Langmuir 31:12894–12902
Banjere RM, Banjere MK, Behera K, Pandey S, Ghosh KK (2020) Micellization behavior of conventional cationic surfactants within glycerol-based deep eutetic solvent. ACS Omega 5:19350–19362
Martínez L, Andrade R, Birgin EG, Martínez JM (2009) Packmol: A package for building initial configurations for molecular dynamics simulations. J Comput Chem 30:2157–2164
Berendsen HJC, van der Spoel D, van Drunen R (1995) GROMACS: A message-passing parallel molecular dynamics implementation. Comp Phys Comm 91:43–56
Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindahl E (2015) GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. Software X 1:19–25
Bussi G, Donadio D, Parrinello M (2007) Canonical sampling through velocity rescaling. J Chem Phys 126:014101
Agieienko V, Buchner R (2019) Densities, Viscosities, and Electrical Conductivities of Pure Anhydrous Reline and Its Mixtures with Water in the Temperature Range (293.15 to 338.15) K. J Chem Eng Data 64:4763–4774
Berendsen HJC, Postma JPM, DiNola A, Haak JR (1984) Molecular dynamics with coupling to an external bath. J Chem Phys 81:3684–3690
Jorgensen WL, Maxwell DS, Tirado-Rives J (1996) Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J Am Chem Soc 118:11225–11236
Granovsky AA Firefly (formerly PC Gamess http://classic.chem.msu.su/gran/firefly/index.html. 2009) homepage
Schmidt MW, Baldridge KK, Boatz JA, Elbert ST, Gordon MS, Jensen JH; Koseki S, Matsunaga N, Nguyen KA, Su S, Windus TL, Dupuis M, Montgomery JA (1993) General atomic and molecular electronic structure system. J Comput Chem 14:1347-1363
de Souza ÍFT, Paschoal VH, Bernardino K, Lima TA, Daemen LL, Ribeiro MCC (2021) Vibrational spectroscopy and molecular dynamics simulation of choline oxyanions salts. J Mol Liq 340:117100
Neese F (2012) The ORCA program system. Wiley Interd. Rev.-Comput Mol, Science 2:73–78
Berendsen HJC, Postma JPM, van Gunsteren WF, Hermans J (1981) Interaction models for water in relation to protein hydration. In: Pullman B (ed) Intermolecular Forces, Dordrecht: D. Dordrecht: D. Reidel Publishing Company, Boston, pp 331–342
Åqvist J (1990) Ion-water interaction potentials derived from free energy perturbation simulations. J Phys Chem 94:8021–8024
Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG (1995) A smooth particle mesh Ewald method. J Chem Phys 103:8577–8593
Humphrey W, Dalke A, Schulten K (1996) VMD - Visual molecular dynamics. J Mol Graphics 14(1):33–38
de Moura AF, Bernardino K, de Oliveira OV, Freitas LCG (2011) Solvation of Sodium Octanoate Micelles in Concentrated Urea Solution Studied by Means of Molecular Dynamics Simulations. J Phys Chem B 115:14582–14590
Dill KA, Bromberg S (2002) Molecular Driving Forces : Statistical Thermodynamics in Chemistry and Biology. Garland Science, New York
Acknowledgements
We acknowledge the financial support from FAPESP (Grant Numbers: 2022/15862-7 and 2023/09350-6) and the “Laboratório Nacional de Computação Científica (LNCC/MCTI, Brazil)” for the use of the supercomputer SDumont (https://sdumont.lncc.br).
Funding
Financial support was provided by the FAPESP (Grant Numbers 2022/15862–7 and 2023/09350–6) and the “Laboratório Nacional de Computação Científica (LNCC/MCTI, Brazil)” for the use of the supercomputer SDumont (https://sdumont.lncc.br).
Author information
Authors and Affiliations
Contributions
R.E. Performed the simulations and analyses, figures and tables preparation K.B. Conceptualization, project administration, supervision, funding acquisition Both authors contributed to the writing and revision of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Eliasquevici, R., Bernardino, K. Counter-ion adsorption and electrostatic potential in sodium and choline dodecyl sulfate micelles — a molecular dynamics simulation study. J Mol Model 30, 101 (2024). https://doi.org/10.1007/s00894-024-05897-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-024-05897-1