Abstract
Context
The specialised family of triggering receptors expressed on myeloid cells (TREMs) plays a pivotal role in causing neurodegenerative disorders and activating microglial anti-inflammatory responses. Nasu-Hakola disease (NHD), a rare autosomal recessive disorder, has been associated with mutations in TREM2, which is also responsible for raising the risk of Alzheimer’s disease (AD). Herein, we have made an endeavour to differentiate the confirmed pathogenic variants in TREM2 extra-cellular domain (ECD) linked with NHD and AD using mutation-induced fold stability change (∆∆G), with the computation of 12distinct structure-based methods through saturation mutagenesis. Correlation analysis between relative solvent accessibility (RSA) and ∆∆G expresses the discrete distributive behaviour of mutants associated with TREM2 in AD (R2 = 0.061) and NHD (R2 = 0.601). Our findings put an emphasis on W50 and V126 as major players in maintaining V-like domain in TREM2. Interestingly, we discern that both of them interact with a common residue Y108, which is dissolved upon mutation. This Y108 could have structural or functional role for TREM2 which can be an ideal candidate for further study. Furthermore, the residual interaction network highlights the importance of R47 and R62 in maintaining the CDR loops that are crucial for ligand binding. Future studies using biophysical characterisation of ligand interactions in TREM2-ECD would be helpful for the development of novel therapeutics for AD and NHD.
Methods
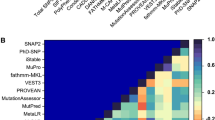
ConSurf algorithm and ENDscript were used to determine the position and conservation of each residue in the wild-type ECD of TREM2. The mutation-induced fold stability change (∆∆G) of confirmed pathogenic mutants associated with NHD and AD was estimated using 12 state-of-the-art structure-based protein stability tools. Furthermore, we also computed the effect of random mutation on these sites using computational saturation mutagenesis. Linear regression analysis was performed using mutants ∆∆G and RSA through GraphPad software. In addition, a comprehensive non-bonded residual interaction network (RIN) of wild type and its mutants of TREM2-ECD was enumerated using RING3.0.





Similar content being viewed by others
Abbreviations
- AD:
-
Alzheimer’s disease
- Aβ:
-
Amyloid beta
- CDR:
-
Complementarity-determining regions
- ECD:
-
Extra-cellular domain
- NHD:
-
Nasu-Hakola disease
- RIN:
-
Residue interaction network
- RSA:
-
Relative solvent accessibility
- TREM:
-
Triggering receptors expressed on myeloid
References
Takahashi K, Rochford CDP, Neumann H (2005) Clearance of apoptotic neurons without inflammation by microglial triggering receptor expressed on myeloid cells-2. J Exp Med 201:647–657. https://doi.org/10.1084/jem.20041611
Bouchon A, Dietrich J, Colonna M (2000) Cutting edge: inflammatory responses can be triggered by TREM-1, a novel receptor expressed on neutrophils and monocytes. J Immunol 164:4991–4995. https://doi.org/10.4049/jimmunol.164.10.4991
Allcock RJN, Barrow AD, Forbes S, Beck S, Trowsdale J (2003) The human TREM gene cluster at 6p21.1 encodes both activating and inhibitory single lgV domain receptors and includes NKp44. Eur J Immunol 33:567–577. https://doi.org/10.1002/immu.200310033
Gratuze M, Leyns CEG, Holtzman DM (2018) New insights into the role of TREM2 in Alzheimer’s disease. Mol Neurodegener 13. https://doi.org/10.1186/s13024-018-0298-9
Borroni B, Ferrari F, Galimberti D, Nacmias B, Barone C, Bagnoli S, Fenoglio C, Piaceri I, Archetti S, Bonvicini C, Gennarelli M, Turla M, Scarpini E, Sorbi S, Padovani A (2014) Heterozygous TREM2 mutations in frontotemporal dementia. Neurobiol Aging 35(934):e7-934.e10. https://doi.org/10.1016/j.neurobiolaging.2013.09.017
Lue LF, Schmitz CT, Serrano G, Sue LI, Beach TG, Walker DG (2015) TREM2 protein expression changes correlate with Alzheimer’s disease neurodegenerative pathologies in post-mortem temporal cortices. Brain Pathol 25:469–480. https://doi.org/10.1111/bpa.12190
Koth LL, Cambier CJ, Ellwanger A, Solon M, Hou L, Lanier LL, Abram CL, Hamerman JA, Woodruff PG (2010) DAP12 is required for macrophage recruitment to the lung in response to cigarette smoke and chemotaxis toward CCL2. J Immunol 184:6522–6528. https://doi.org/10.4049/jimmunol.0901171
Wang Y, Ulland TK, Ulrich JD, Song W, Tzaferis JA, Hole JT, Yuan P, Mahan TE, Shi Y, Gilfillan S, Cella M, Grutzendler J, DeMattos RB, Cirrito JR, Holtzman DM, Colonna M (2016) TREM2-mediated early microglial response limits diffusion and toxicity of amyloid plaques. J Exp Med 213:667–675. https://doi.org/10.1084/jem.20151948
Otero K, Shinohara M, Zhao H, Cella M, Gilfillan S, Colucci A, Faccio R, Ross FP, Teitelbaum SL, Takayanagi H, Colonna M (2012) TREM2 and β-catenin regulate bone homeostasis by controlling the rate of osteoclastogenesis. J Immunol 188:2612–2621. https://doi.org/10.4049/jimmunol.1102836
Carlyle WC, Mcclain JB, Tzafriri AR, Bailey L, Brett G, Markham PM, Stanley JRL, Edelman ER, Sciences CB, Technologies E, Park LT (2015) TREM2 maintains microglial metabolic fitness in Alzheimer’s disease. Cell 162:561–567
Andreone BJ, Przybyla L, Llapashtica C, Rana A, Davis SS, van Lengerich B, Lin K, Shi J, Mei Y, Astarita G, Di Paolo G, Sandmann T, Monroe KM, Lewcock JW (2020) Alzheimer’s-associated PLCγ2 is a signaling node required for both TREM2 function and the inflammatory response in human microglia. Nat Neurosci 23:927–938. https://doi.org/10.1038/s41593-020-0650-6
Bouchon A, Hernández-Munain C, Cella M, Colonna M (2001) A DAP12-mediated pathway regulates expression of CC chemokine receptor 7 and maturation of human dendritic cells. J Exp Med 194:1111–1122. https://doi.org/10.1084/jem.194.8.1111
Kawabori M, Kacimi R, Kauppinen T, Calosing C, Kim JY, Hsieh CL, Nakamura MC, Yenari MA (2015) Triggering receptor expressed on myeloid cells 2 (TREM2) deficiency attenuates phagocytic activities of microglia and exacerbates ischemic damage in experimental stroke. J Neurosci 35:3384–3396. https://doi.org/10.1523/JNEUROSCI.2620-14.2015
Hakola HP (1972) Neuropsychiatric and genetic aspects of a new hereditary disease characterized by progressive dementia and lipomembranous polycystic osteodysplasia. Acta Psychiatr Scand Suppl 232:1–173
Lue LF, Schmitz C, Walker DG (2015) What happens to microglial TREM2 in Alzheimer’s disease: immunoregulatory turned into immunopathogenic? Neuroscience 302:138–150. https://doi.org/10.1016/j.neuroscience.2014.09.050
Deczkowska A, Weiner A, Amit I (2020) The physiology, pathology, and potential therapeutic applications of the TREM2 signaling pathway. Cell 181:1207–1217. https://doi.org/10.1016/j.cell.2020.05.003
Wunderlich P, Glebov K, Kemmerling N, Tien NT, Neumann H, Walter J (2013) Sequential proteolytic processing of the triggering receptor expressed on myeloid cells-2 (TREM2) protein by ectodomain shedding and γ-secretase- dependent intramembranous cleavage. J Biol Chem 288:33027–33036. https://doi.org/10.1074/jbc.M113.517540
Takalo M, Wittrahm R, Wefers B, Parhizkar S, Jokivarsi K, Kuulasmaa T, Mäkinen P, Martiskainen H, Wurst W, Xiang X, Marttinen M, Poutiainen P, Haapasalo A, Hiltunen M, Haass C (2020) The Alzheimer’s disease-associated protective Plcγ2-P522R variant promotes immune functions. Mol Neurodegener 15. https://doi.org/10.1186/s13024-020-00402-7
Molinuevo JL, Ayton S, Batrla R, Bednar MM, Bittner T, Cummings J, Fagan AM, Hampel H, Mielke MM, Mikulskis A, O’bryant S, Scheltens P, Sevigny J, Shaw LM, Soares HD, Tong G, Trojanowski JQ, Zetterberg H, Blennow K (2018) Current state of Alzheimer’s fluid biomarkers. Acta Neuropathol. 136:821–853. https://doi.org/10.1007/s00401-018-1932-x
Filipello F, Goldsbury C, You SF, Locca A, Karch CM, Piccio L (2022) Soluble TREM2: innocent bystander or active player in neurological diseases? Neurobiol Dis 165:105630. https://doi.org/10.1016/j.nbd.2022.105630
Suárez‐Calvet M, Kleinberger G, Araque Caballero MÁ, Brendel M, Rominger A, Alcolea D, Fortea J, Lleó A, Blesa R, Gispert JD, Sánchez‐Valle R (2016) sTREM-2 cerebrospinal fluid levels are a potential biomarker for microglia activity in early-stage Alzheimer's disease and associate with neuronal injury markers. EMBO Mol Med 8: 466–476. https://doi.org/10.15252/emmm.201506123
Rodriguez-Vieitez E, Ashton NJ (2021) Plasma sTREM2: a potential marker of cerebrovascular injury in neurodegenerative disorders. Brain 144:3283–3285. https://doi.org/10.1093/brain/awab399
Guerreiro R, Wojtas A, Bras J, Carrasquillo M, Rogaeva E, Majounie E, Cruchaga C, Sassi C, Kauwe JSK, Younkin S, Hazrati L, Collinge J, Pocock J, Lashley T, Williams J, Lambert J-C, Amouyel P, Goate A, Rademakers R, Morgan K, Powell J, St. George-Hyslop P, Singleton A, Hardy J (2013) TREM2 variants in Alzheimer’s disease. N Engl J Med 368:117–127. https://doi.org/10.1056/nejmoa1211851
Jonsson T, Stefansson H, Steinberg S, Jonsdottir I, Jonsson PV, Snaedal J, Bjornsson S, Huttenlocher J, Levey AI, Lah JJ, Rujescu D (2013) Variant of TREM2 associated with the risk of Alzheimer’s disease. N Engl J Med 368:107–116. https://doi.org/10.1056/NEJMoa1211103
Rayaprolu S, Mullen B, Baker M, Lynch T, Finger E, Seeley WW, Hatanpaa KJ, Lomen-Hoerth C, Kertesz A, Bigio EH, Lippa C (2013) TREM2 in neurodegeneration: evidence for association of the p. R47H variant with frontotemporal dementia and Parkinson’s disease. Mol Neurodegener 8:1–5. https://doi.org/10.1186/1750-1326-8-19
Cuyvers E, Bettens K, Philtjens S, Van Langenhove T, Gijselinck I, van der Zee J, Engelborghs S, Vandenbulcke M, Van Dongen J, Geerts N, Maes G, Mattheijssens M, Peeters K, Cras P, Vandenberghe R, De Deyn PP, Van Broeckhoven C, Cruts M, Sleegers K (2014) Investigating the role of rare heterozygous TREM2 variants in Alzheimer’s disease and frontotemporal dementia. Neurobiol Aging 35(726):e11-726.e19. https://doi.org/10.1016/j.neurobiolaging.2013.09.009
Holtzman DM, Morris JC, Goate AM (2011) Alzheimer’s disease: the challenge of the second century. Sci Transl Med 3. https://doi.org/10.1126/scitranslmed.3002369
Hardy J, Selkoe DJ (2002) The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science (80-. ) 297:353–356. https://doi.org/10.1126/science.1072994
Area-Gomez E, Schon EA (2017) Alzheimer disease. Adv Exp Med Biol 997:149–156. https://doi.org/10.1007/978-981-10-4567-7_11
Huang et al (2002) Alzheimer mechanisms and therapeutic strategies. Science (80-. ) 148:1204–1222
Schoepp NG, Khorosheva EM, Schlappi TS, Curtis MS, Humphries RM, Angeles L, Annex B, Angeles L, Hindler JA, Angeles L, Annex B, Angeles L, Ismagilov RF (2017) TREM2 haplodeficiency in mice and humans impairs the microglia barrier function leading to decreased amyloid compaction and severe axonal dystrophy. Neuron 55:9557–9561
Keren-Shaul H, Spinrad A, Weiner A, Matcovitch-Natan O, Dvir-Szternfeld R, Ulland TK, David E, Baruch K, Lara-Astaiso D, Toth B, Itzkovitz S, Colonna M, Schwartz M, Amit I (2017) A unique microglia type associated with restricting development of Alzheimer’s disease. Cell 169:1276-1290.e17. https://doi.org/10.1016/j.cell.2017.05.018
Condello C, Yuan P, Schain A, Grutzendler J (2015) Microglia constitute a barrier that prevents neurotoxic protofibrillar Aβ42 hotspots around plaques. Nat Commun 6. https://doi.org/10.1038/ncomms7176
Colonna M, Wang Y (2016) TREM2 variants: new keys to decipher Alzheimer disease pathogenesis. Nat Rev Neurosci 17:201–207. https://doi.org/10.1038/nrn.2016.7
Sudom A, Talreja S, Danao J, Bragg E, Kegel R, Min X, Richardson J, Zhang Z, Sharkov N, Marcora E, Thibault S, Bradley J, Wood S, Lim AC, Chen H, Wang S, Foltz IN, Sambashivan S, Wang Z (2018) Molecular basis for the loss-of-function effects of the Alzheimer’s disease-associated R47H variant of the immune receptor TREM2. J Biol Chem 293:12634–12646. https://doi.org/10.1074/jbc.RA118.00235
Cheng-Hathaway PJ, Reed-Geaghan EG, Jay TR, Casali BT, Bemiller SM, Puntambekar SS, Von Saucken VE, Williams RY, Karlo JC, Moutinho M, Xu G, Ransohoff RM, Lamb BT, Landreth GE (2018) The Trem2 R47H variant confers loss-of-function-like phenotypes in Alzheimer’s disease. Mol Neurodegener 13. https://doi.org/10.1186/s13024-018-0262-8
Luis EO, Ortega-Cubero S, Lamet I, Razquin C, Cruchaga C, Benitez BA, Lorenzo E, Irigoyen J, Pastor MA, Pastor P, Initiative Alzheimer’s Disease Neuroimaging, (ADNI), (2014) Frontobasal gray matter loss is associated with the TREM2 p. R47H variant. Neurobiol Aging 35:2681–2690. https://doi.org/10.1016/j.neurobiolaging.2014.06.007
Del-Aguila JL, Fernández MV, Schindler S, Ibanez L, Deming Y, Ma S, Saef B, Black K, Budde J, Norton J, Chasse R, Harari O, Goate A, Xiong C, Morris JC, Cruchaga C (2018) Assessment of the genetic architecture of Alzheimer’s disease risk in rate of memory decline. J Alzheimer’s Dis 62:745–756. https://doi.org/10.3233/JAD-170834
Jin SC, Benitez BA, Karch CM, Cooper B, Skorupa T, Carrell D, Norton JB, Hsu S, Harari O, Cai Y, Bertelsen S, Goate AM, Cruchaga C (2014) Coding variants in TREM2 increase risk for Alzheimer’s disease. Hum Mol Genet 23:5838–5846. https://doi.org/10.1093/hmg/ddu277
Sims R, Badarinarayan N, Raybould R, Heilmann-Heimbach S, Vronskaya M, Hoffmann P (2017) Rare coding variants in PLCG2, ABI3 and TREM2 implicate microglial-mediated innate immunity in Alzheimer’s disease. Nat Genet 49:1373–1384
Jiang T, Hou J-K, Gao Q, Yu J-T, Zhou J-S, Zhao H-D, Zhang Y-D (2016) TREM2 p.H157Y variant and the risk of Alzheimer’s disease: a meta-analysis involving 14,510 subjects. Curr Neurovasc Res 13:318–320. https://doi.org/10.2174/1567202613666160808095530
Paloneva J, Kestilä M, Wu J, Salminen A, Böhling T, Ruotsalainen V, Hakola P, Bakker AB, Phillips JH, Pekkarinen P, Lanier LL (2000) Loss-of-function mutations in TYROBP (DAP12) result in a presenile dementia with bone cysts. Nat Genet 25:357–361. https://doi.org/10.1038/77153
Paloneva J, Manninen T, Christman G, Hovanes K, Mandelin J, Adolfsson R, Bianchin M, Bird T, Miranda R, Salmaggi A, Tranebjærg L, Konttinen Y, Peltonen L (2002) Mutations in two genes encoding different subunits of a receptor signaling complex result in an identical disease phenotype. Am J Hum Genet 71:656–662. https://doi.org/10.1086/342259
Dardiotis E, Siokas V, Pantazi E, Dardioti M, Rikos D, Xiromerisiou G, Markou A, Papadimitriou D, Speletas M, Hadjigeorgiou GM (2017) A novel mutation in TREM2 gene causing Nasu-Hakola disease and review of the literature. Neurobiol Aging 53(194):e13-194.e22. https://doi.org/10.1016/j.neurobiolaging.2017.01.015
Klünemann HH, Ridha BH, Magy L, Wherrett JR, Hemelsoet DM, Keen RW, De Bleecker JL, Rossor MN, Marienhagen J, Klein HE, Peltonen L, Paloneva J (2005) The genetic causes of basal ganglia calcification, dementia, and bone cysts: DAP12 and TREM2. Neurology 64:1502–1507. https://doi.org/10.1212/01.WNL.0000160304.00003.CA
Paloneva J, Autti T, Raininko R, Partanen J, Salonen O, Puranen M, Hakola P, Haltia M (2001) CNS manifestations of Nasu-Hakola disease: a frontal dementia with bone cysts. Neurology 56:1552–1558. https://doi.org/10.1212/wnl.56.11.1552
Kober DL, Alexander-Brett JM, Karch CM, Cruchaga C, Colonna M, Holtzman MJ, Brett TJ (2016) Neurodegenerative disease mutations in TREM2 reveal a functional surface and distinct loss-of-function mechanisms. Elife 5. https://doi.org/10.7554/eLife.20391
Sirkis DW, Aparicio RE, Schekman R (2017) Neurodegeneration-associated mutant TREM2 proteins abortively cycle between the ER and ER–Golgi intermediate compartment. Mol Biol Cell 28:2723–2733. https://doi.org/10.1091/mbc.e17-06-0423
Paloneva J, Mandelin J, Kiialainen A, Böhling T, Prudlo J, Hakola P, Haltia M, Konttinen YT, Peltonen L (2003) DAP12/TREM2 deficiency results in impaired osteoclast differentiation and osteoporotic features. J Exp Med 198:669–675. https://doi.org/10.1084/jem.20030027
Song W, Hooli B, Mullin K, Jin SC, Cella M, Ulland TK, Wang Y, Tanzi RE, Colonna M (2017) Alzheimer’s disease-associated TREM2 variants exhibit either decreased or increased ligand-dependent activation. Alzheimer’s Dement 13:381–387. https://doi.org/10.1016/j.jalz.2016.07.004
Tang N, Dehury B, Kepp KP (2019) Computing the pathogenicity of Alzheimer’s disease presenilin 1 mutations. J Chem Inf Model 59:858–870. https://doi.org/10.1021/acs.jcim.8b00896
Berman PEBHM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The Protein Data Bank nucleic acids research, 28: 235–242. Protein Data Bank Nucleic Acids Res 28(2000):235–242
Ashkenazy H, Abadi S, Martz E, Chay O, Mayrose I, Pupko T, Ben-Tal N (2016) ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res 44(2016):W344–W350. https://doi.org/10.1093/NAR/GKW408
Gouet P, Robert X, Courcelle E (2003) ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res 31:3320–3323. https://doi.org/10.1093/nar/gkg556
Schrödinger L (n.d.) The PyMOL Molecular Graphics, Version 2.5.2
Daggett V, Fersht AR (2003) Is there a unifying mechanism for protein folding? Trends Biochem Sci 28:18–25. https://doi.org/10.1016/S0968-0004(02)00012-9
Gromiha MM, Huang L-T (2011) Machine learning algorithms for predicting protein folding rates and stability of mutant proteins: comparison with statistical methods. Curr Protein Pept Sci 12:490–502. https://doi.org/10.2174/138920311796957630
Dehouck Y, Kwasigroch JM, Gilis D, Rooman M (2011) PoPMuSiC 2.1: a web server for the estimation of protein stability changes upon mutation and sequence optimality. BMC Bioinformatics 12. https://doi.org/10.1186/1471-2105-12-151
Parthiban V, Gromiha MM, Schomburg D (2006) CUPSAT: prediction of protein stability upon point mutations. Nucleic Acids Res 34. https://doi.org/10.1093/nar/gkl190
Pires DEV, Ascher DB, Blundell TL (2014) MCSM: predicting the effects of mutations in proteins using graph-based signatures. Bioinformatics 30:335–342. https://doi.org/10.1093/bioinformatics/btt691
Capriotti E, Fariselli P, Casadio R (2005) I-Mutant2.0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res 33. https://doi.org/10.1093/nar/gki375
Musil M, Stourac J, Bendl J, Brezovsky J, Prokop Z, Zendulka J, Martinek T, Bednar D, Damborsky J (2017) FireProt: web server for automated design of thermostable proteins. Nucleic Acids Res 45:W393–W399. https://doi.org/10.1093/nar/gkx285
Schymkowitz J, Borg J, Stricher F, Nys R, Rousseau F, Serrano L (2005) The FoldX web server: an online force field. Nucleic Acids Research 33:W382–W388. https://doi.org/10.1093/nar/gki387
Giollo M, Martin AJM, Walsh I, Ferrari C, Tosatto SCE (2014) NeEMO: a method using residue interaction networks to improve prediction of protein stability upon mutation. BMC Genomics 15:1–11. https://doi.org/10.1186/1471-2164-15-S4-S7
Pucci F, Bourgeas R, Rooman M (2016) Predicting protein thermal stability changes upon point mutations using statistical potentials: introducing HoTMuSiC. Sci Rep 6. https://doi.org/10.1038/srep23257
Worth CL, Preissner R, Blundell TL (2011) SDM - a server for predicting effects of mutations on protein stability and malfunction. Nucleic Acids Res 39. https://doi.org/10.1093/nar/gkr363
Pires DEV, Ascher DB, Blundell TL (2014) DUET: a server for predicting effects of mutations on protein stability using an integrated computational approach. Nucleic Acids Res 42. https://doi.org/10.1093/nar/gku411
Rodrigues CHM, Pires DEV, Ascher DB (2018) DynaMut: predicting the impact of mutations on protein conformation, flexibility and stability. Nucleic Acids Res 46:W350–W355. https://doi.org/10.1093/nar/gky300
Chen Y, Lu H, Zhang N, Zhu Z, Wang S, Li M (2020) PremPS: predicting the impact of missense mutations on protein stability. PLoS Comput Biol 16:1–22. https://doi.org/10.1371/journal.pcbi.1008543
Montanucci L, Capriotti E, Birolo G, Benevenuta S, Pancotti C, Lal D, Fariselli P (2022) DDGun: an untrained predictor of protein stability changes upon amino acid variants. Nucleic Acids Res 50:W222–W227. https://doi.org/10.1093/nar/gkac325
Clementel D, Del Conte A, Monzon AM, Camagni GF, Minervini G, Piovesan D, Tosatto SCE (2022) RING 3.0: fast generation of probabilistic residue interaction networks from structural ensembles. Nucleic Acids Res. 50:W651–W656. https://doi.org/10.1093/nar/gkac365
Li Z, Del-Aguila JL, Dube U, Budde J, Martinez R, Black K, Xiao Q, Cairns NJ, Network The Dominantly Inherited Alzheimer, (DIAN), Dougherty JD, Lee J-M, Morris JC, Bateman RJ, Karch CM, Cruchaga C, Harari O, (2018) Genetic variants associated with Alzheimer’s disease confer different cerebral cortex cell-type population structure. Nucleic Acids Res 10:1–19. https://doi.org/10.1186/s13073-018-0551-4
Sudom A, Talreja S, Danao J, Bragg E, Kegel R, Min X, Richardson J, Zhang Z, Sharkov N, Marcora E, Thibault S, Bradley J, Wood S, Lim A-C, Chen H, Wang S, Foltz IN, Sambashivan S, Wang Z (2018) Molecular basis for the loss-of-function effects of the Alzheimer’s disease–associated R47H variant of the immune receptor TREM2. J Biol Chem 293:12634–12646. https://doi.org/10.1074/jbc.ra118.002352
Dash R, Choi HJ, Moon IS (2020) Mechanistic insights into the deleterious roles of Nasu-Hakola disease associated TREM2 variants. Sci Rep 10. https://doi.org/10.1038/s41598-020-60561-x
Guerreiro RJ, Lohmann E, Brás JM, Gibbs JR, Rohrer JD, Gurunlian N, Dursun B, Bilgic B, Hanagasi H, Gurvit H, Emre M, Singleton A, Hardy J (2012) Using exome sequencing to reveal mutations in TREM2 presenting as a frontotemporal dementia–like syndrome without bone involvement. JAMA Neurol 1. https://doi.org/10.1001/archneurol.2013.579
Ahmad F (2022) Protein stability [determination] problems. Front Mol Biosci 9. https://doi.org/10.3389/fmolb.2022.880358
Bommarius AS, Paye MF (2013) Stabilizing biocatalysts. Chem Soc Rev 42:6534–6565. https://doi.org/10.1039/c3cs60137d
Magliery TJ (2015) Protein stability: computation, sequence statistics, and new experimental methods. Curr Opin Struct Biol 33:161–168. https://doi.org/10.1016/j.sbi.2015.09.002
Sauerborn M, Brinks V, Jiskoot W, Schellekens H (2010) Immunological mechanism underlying the immune response to recombinant human protein therapeutics. Trends Pharmacol Sci 31:53–59. https://doi.org/10.1016/j.tips.2009.11.001
Manning MC, Chou DK, Murphy BM, Payne RW, Katayama DS (2010) Stability of protein pharmaceuticals: an update. Pharm Res 27:544–575. https://doi.org/10.1007/s11095-009-0045-6
Boulet-Audet M, Byrne B, Kazarian SG (2014) High-throughput thermal stability analysis of a monoclonal antibody by attenuated total reflection FT-IR spectroscopic imaging. Anal Chem 86:9786–9793. https://doi.org/10.1021/ac502529q
Uversky V, Breydo L, Redington J (2017) When good goes awry: the aggregation of protein therapeutics. Protein Pept Lett 24:340–347. https://doi.org/10.2174/0929866524666170209153421
Bloom JD, Labthavikul ST, Otey CR, Arnold FH (2006) Protein stability promotes evolvability. Proc Natl Acad Sci U S A 103:5869–5874. https://doi.org/10.1073/pnas.0510098103
Nobuhiko T, Tawfik DS (2009) Stability effects of mutations and protein evolvability. Curr Opin Struct Biol 19:596–604
Reetz MT, Carballeira JD (2007) Iterative saturation mutagenesis (ISM) for rapid directed evolution of functional enzymes. Nat Protoc 2:891–903. https://doi.org/10.1038/nprot.2007.72
Stourac J, Dubrava J, Musil M, Horackova J, Damborsky J, Mazurenko S, Bednar D (2021) FireProtDB: database of manually curated protein stability data. Nucleic Acids Res 49:D319–D324. https://doi.org/10.1093/nar/gkaa981
Kellogg EH, Leaver-Fay A, Baker D (2011) Role of conformational sampling in computing mutation-induced changes in protein structure and stability. Proteins Struct Funct Bioinforma 79:830–838. https://doi.org/10.1002/prot.22921
Reifschneider A, Robinson S, van Lengerich B, Gnörich J, Logan T, Heindl S, Vogt MA, Weidinger E, Riedl L, Wind K, Zatcepin A, Pesämaa I, Haberl S, Nuscher B, Kleinberger G, Klimmt J, Götzl JK, Liesz A, Bürger K, Brendel M, Levin J, Diehl‐Schmid J, Suh J, Di Paolo G, Lewcock JW, Monroe KM, Paquet D, Capell A, Haass C (2022) Loss of TREM2 rescues hyperactivation of microglia, but not lysosomal deficits and neurotoxicity in models of progranulin deficiency. EMBO J 41. https://doi.org/10.15252/embj.2021109108
Dash R, Munni YA, Mitra S, Choi HJ, Jahan SI, Chowdhury A, Jang TJ, Moon IS (2022) Dynamic insights into the effects of nonsynonymous polymorphisms (nsSNPs) on loss of TREM2 function. Sci Rep 12. https://doi.org/10.1038/s41598-022-13120-5
Hall-Roberts H, Agarwal D, Obst J, Smith TB, Monzón-Sandoval J, Di Daniel E, Webber C, James WS, Mead E, Davis JB, Cowley SA (2020) TREM2 Alzheimer’s variant R47H causes similar transcriptional dysregulation to knockout, yet only subtle functional phenotypes in human iPSC-derived macrophages. Alzheimer’s Res Ther 12:1–27. https://doi.org/10.1186/s13195-020-00709-z
Yeh FL, Wang Y, Tom I, Gonzalez LC, Sheng M (2016) TREM2 binds to apolipoproteins, including APOE and CLU/APOJ, and thereby facilitates uptake of amyloid-beta by microglia. Neuron 91:328–340. https://doi.org/10.1016/j.neuron.2016.06.015
Patel D, Mez J, Vardarajan BN, Staley L, Chung J, Zhang X, Farrell JJ, Rynkiewicz MJ, Cannon-Albright LA, Teerlink CC, Stevens J, Corcoran C, Gonzalez Murcia JD, Lopez OL, Mayeux R, Haines JL, Pericak-Vance MA, Schellenberg G, Kauwe JSK, Lunetta KL, Farrer LA (2019) Association of rare coding mutations with Alzheimer disease and other dementias among adults of European ancestry. JAMA Netw Open 2(3):e191350. https://doi.org/10.1001/jamanetworkopen.2019.1350
Strub C, Alies C, Lougarre A, Ladurantie C, Czaplicki J, Fournier D (2004) Mutation of exposed hydrophobic amino acids to arginine to increase protein stability. BMC Biochem 5:1–6. https://doi.org/10.1186/1471-2091-5-9
Acknowledgements
The authors acknowledge the support from ICMR-RMRC, Bhubaneswar, for computational facility. The authors also acknowledge DHR for providing Young Scientist grant to BD. The authors also acknowledge ICMR for providing ICMR-Centenary Postdoctoral Fellowship to SP.
Author information
Authors and Affiliations
Contributions
Preety Sthutika Swain: methodology, software investigation, data curation, visualisation, writing — original draft.
Sunita Panda: data curation, methodology, review and editing.
Sanghamitra Pati: writing — review and editing.
Budheswar Dehury: conceptualisation, methodology, review and editing, and supervision.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
The data supporting this manuscript is listed in the “Supplementary information” section.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Swain, P.S., Panda, S., Pati, S. et al. Computational saturation mutagenesis to explore the effect of pathogenic mutations on extra-cellular domains of TREM2 associated with Alzheimer’s and Nasu-Hakola disease. J Mol Model 29, 360 (2023). https://doi.org/10.1007/s00894-023-05770-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-023-05770-7