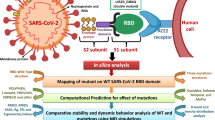
Abstract
Objective
The COVID-19 epidemic is raging around the world, with the emergence of viral mutant strains such as Delta and Omicron, posing severe challenges to people’s health and quality of life. A full understanding life cycle of the virus in host cells helps to reveal inactivation mechanism of antibody and provide inspiration for the development of a new-generation vaccines.
Methods
In this work, molecular recognitions and conformational changes of SARS-CoV-2 spike protein mutants (i.e., Delta, Mu, and Omicron) and three essential partners (i.e., membrane receptor hACE2, protease TMPRSS2, and antibody C121) both were compared and analyzed using molecular simulations.
Results
Water basin and binding free energy calculations both show that the three mutants possess higher affinity for hACE2 than WT, exhibiting stronger virus transmission. The descending order of cleavage ability by TMPRSS2 is Mu, Delta, Omicron, and WT, which is related to the new S1/S2 cutting site induced by transposition effect. The inefficient utilization of TMPRSS2 by Omicron is consistent with its primary entry into cells via the endosomal pathway. In addition, RBD-directed antibody C121 showed obvious resistance to Omicron, which may have originated from high fluctuation of approaching angles, high flexibility of I472-F490 loop, and reduced binding ability.
Conclusions
According to the overall characteristics of the three mutants, high infectivity, high immune escape, and low virulence may be the future evolutionary selection of SARS-CoV-2. In a word, this work not only proposes the possible resistance mechanism of SARS-CoV-2 mutants, but also provides theoretical guidance for the subsequent drug design against COVID-19 based on S protein structure.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
As the largest RNA virus, coronaviruses contain about 30 kb of a non-segmental positive-sense genome, which can cause different diseases in poultry and mammals. The current three coronavirus causing human disease are all β-CoVs, including the severe acute respiratory syndrome coronavirus (SARS-CoV), the Middle East respiratory syndrome coronavirus (MERS-CoV), and the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [1]. SARS-CoV was initially reported in Guangdong China in 2002 [2], and infected over 8000 people worldwide with fatality rate of 10% [3, 4]. MERS-CoV emerged in Saudi Arabia in June 2012 [5], showing low infection rate but higher fatality rate of 34.4% [1]. On February 11, 2020, the acute pneumonia caused by SARS-CoV-2 has been named COVID-19. It has led to over 500 million infections and 6 million deaths worldwide due to its rapid global spread, long incubation period, and proclivity to evolve new variants [6]. Rabaan et al. revealed that SARS-CoV-2 is structurally similar to SARS-CoV in general, but slightly different in spike protein [7]—the most important structural protein [8]. Compared with SARS and MERS, SARS-CoV-2 is less pathogenic, while possessing higher transmission rate in humans due to the presence of a furin-like cleavage site in spike and high affinity with human angiotensin-converting enzyme 2 (hACE2) receptor [8, 9].
For searching an effective therapy, it is necessary to understand the transmission of SARS-CoV-2 between host populations, especially multibody interactions of key viral proteins including spike. The SARS-CoV-2 spike consists of S1 and S2 subunits: the former comprises a receptor binding domain (RBD) to initiate viral entry into the target cell, as well as an N-terminal domain (NTD); [10] the latter contains six domains (i.e., fusion peptide, HR1 (heptapeptide repeat 1), central helix, HR2 (heptapeptide repeat 2) junction domain, transmembrane domain, and cytoplasmic tail), which mediate the fusion of viral to host cellular membranes after significant conformational rearrangements [1, 11,12,13,14]. The S protein binds to hACE2 via RBD, which is a key step of membrane fusion and is closely related to the infection, tropism, and pathogenicity of SARS-CoV-2 [8, 15, 16]. The up- and down-conformations of spike were observed prior to membrane fusion, in which the up-state was more conducive to molecular recognition by hACE2 receptor [17,18,19]. S protein is then cleaved and activated by furin-like protease and transmembrane protease serine 2 (TMPRSS2) in turn, undergoing significant structural rearrangements [20,21,22,23,24]. After membrane fusion and endocytosis mediated by transmembrane proteases, the genomic RNA is eventually expressed and replicated to assemble new coronaviruses particles. In sum, RBD conformational state of spike, the interactions respectively with hACE2 and TMPRSS2 are all key scientific questions to reveal the life cycle of SARS-CoV-2 [25].
In the face of the COVID-19 pandemic, the structure and transmission of the pathogen SARS-CoV-2 have received the greatest attention, and numerous effective intervention measures have emerged successively. RNA vaccines, live attenuated vaccines, and inactivated vaccines are all extensively used around the world as the primary means of prevention and treatment against COVID-19. In addition to them, there are several types of vaccines, all of which are in phases 3 or 4 clinical trials (see Fig. 1) [26]. By April 11, 2022, total 344 SARS-CoV-2 vaccine candidates have been developed and tested in various stages of clinical trials, and 31 of them in use [27]. Universal vaccination appears to prevent the severe course of illness and deaths caused by all occurring variants of concern (VOCs) [28, 29]. By April 13, 2022, a total of 11,294,502,059 vaccine doses have been administered all over the world, with 65.5% of the population having received at least one dose of a COVID-19 vaccine [30]. Given the increasing number of infections, booster jab of COVID-19 vaccine had been widely adopted to help enhance the action of antibodies and prolong protection [31, 32]. In addition, some antiviral drugs and therapeutic antibodies are also recommended, such as bamlanivimab, etesevimab [33], sotrovimab, casirivimab, imdevimab [34, 35], Evusheld [36] (acting on RBD to affects the binding with hACE2), and Actemra (inhibiting IL-6 induction) [37]. The previously authorized use of sotrovimab will be discontinued until April 5, 2022, as it may not be effective against the Omicron variant. At present, there are few antiviral drugs, only baricitinib (selective inhibitor against Janus-activated kinases 1 and 2), Paxlovid, and molnupiravir (targeting autophagy and unfolded protein) were reported [38].
After more than 2 years of fighting against the epidemic, the situation remained dire, and many types of variants of concern (VOCs) have emerged, including Delta (B.1.617.2), Mu (B.1.621), and Omicron (B.1.1.529) [39]. The epidemic storm dominated by Delta was popular for a time, showing high infectivity and hospitalization rate [40, 41]. Despite the many existing treatment strategies that have been available, Delta still remained more prevalent than other subtypes in quite a long period of time, with the second highest weekly new cases and moderate weekly deaths (see Fig. 1). The Mu variant was first identified in Colombia in January 2021 [42] and has become a new member of variant of interest (VOI). Subsequently, Mu was shown to be remarkably resistant not only to the sera from COVID-19 convalescents and BNT162b2-vaccinated individuals, but also to the inactivated vaccine-elicited antibodies [43]. On November 26, 2021, the WHO named the new variant B.1.1.529 to be Omicron with a large number of mutations in S protein RBD [44, 45]. Studies on spike VSV pseudovirus have shown that the neutralizing activity against Omicron is substantially reduced after 2 or 3 doses (booster) of inactivated vaccine, falling below the protective threshold in several cases [46, 47]. So far, Omicron has proven to be a highly contagious variant and may evolve new mechanisms infecting humans [47,48,49].
There are a variety of new drugs and treatments for COVID-19 that are still under continuous development. Nevertheless, inadequate vaccination rates and a declining ability to neutralize viral variants both made the situation difficult [50, 51]. In this work, based on molecular simulations of four strains (WT, Delta, Mu, and Omicron) spike. we demonstrated how mutations respectively affect the recognition of S protein with hACE2, TMPRSS2, and C121 (a neutralizing antibody that blocks the association with hACE2) during infection, and then revealed the possible mechanism of vaccine inactivation. It provides theoretical guidance for the development, evaluation, and design of the next generation of vaccines against COVID-19.
Materials and methods
Preparation of simulation systems
The initial S protein structure was taken from the RCSB protein data bank (PDB), with all missing residues completed by the SWISS-MODEL server [52]. Considering that the up-state of spike is necessary for our study the crystal structure of wild-type spike with one RBD up (PDB ID: 7KJ2, bond with hACE2) was selected as the template for subsequent system construction [53]. After deleting the amino acid sequence of deletion mutation and hACE2, the model with deletion mutation was established by SWISS-MODEL. The whole mutants are obtained by completing the residual mutation in the Pymol mutagenesis module [54]. Then, HADDOCK 2.4 sever [55] was used to build the following twelve systems: (1) four spike trimer complexes with hACE2 receptor; (2) four spike trimer docked complex models with TMPRSS2; (3) four spike trimer docked complex models with antibody C121. The structures of TMPRSS2 and C121 were available from PDBs 7MEQ [56] and 7K8X [57], respectively. For convenience of analysis, the twelve SP complex systems are denoted by WT_hACE2/_TMPRSS2/_C121, Delta_hACE2/_TMPRSS2/_C121, Mu_hACE2/_TMPRSS2/_C121, and Omicron_hACE2/_TMPRSS2/_C121, respectively.
Molecular dynamics simulation
Molecular dynamics (MD) simulation has been widely used for exploring the thermodynamic characteristics of biomolecules at the atomic level. In order to analyze how the three drug resistance mutations (i.e., Delta, Mu, and Omicron) affect molecular recognition of S protein and its essential partners, comparative MD simulations were conducted for the twelve S protein complex systems using ff14SB force field and Amber 19 package [58, 59]. All solutes were solvated in octahedral box with the boundary of 15.0 Å by TIP3P water model [60].
To reduce the space collision in simulated systems, two energy optimizations (EM) were carried out. The first EM stage is composed of 5000 steps of steepest descent and 5000 steps of conjugate gradient optimizations with constraint force constant of 500 kcal·mol−1·Å−2; in the second EM stage, the solute constraint is completely removed, also containing the same optimization steps and convergence criterion with energy gradient less than 0.01 kcal·mol−1·Å−2.
After EM procedure, MD simulations were also divided into two stages: (1) 5 ns solute-constrained MD simulations with a force constant of 10 kcal·mol−1 Å−2, where temperature gradually increased from 0 to 300 K; (2) 95 ns solute-unconstrained productive MD simulations, adopting SHAKE algorithm [61] to constrain the hydrogen-containing atoms. During the simulation, non-bonded interaction radius and integration step were set to 10 Å and 2 fs, respectively, and the conformational fluctuation was monitored by VMD 1.9.3 package [62]. The atomic coordinates were stored every 10 ps, and a total of 10,000 conformations were collected for the subsequent analyses of molecular recognition and conformational change.
Binding free energy calculation
As an important physical and chemical parameter, binding free energy can be applied to effectively judge molecular recognition between receptors and ligands, and is also a key criterion for evaluating drug activity. Molecular mechanics/Poisson Boltzmann (MM/PBSA) method was used to calculate the binding free energies between four spike trimers (i.e., the WT, Delta, Mu, and Omicron variants) with hACE2, TMPRSS2, and C121. Total 25 snapshots were collected from each MD trajectory every 4 ns intervals from 1 to 100 ns, and used for the calculation of average binding free energy. The calculation formula is as follows:
where ∆H represents the total enthalpy change, T is the absolute temperature in Kelvin, and ∆S refers to the total entropy change calculated using the normal mode method [63]. ∆EVDW indicates intramolecular van der Waals energy under vacuum, while ∆EELE refers to the electrostatic fraction. ∆GPBELE and ∆GPBSUR represent the hydrophilic and hydrophobic parts of solvation binding free energy, respectively.
Identification of key residues
Energy decomposition was carried out by molecular mechanics/generalized Born area (MM/GBSA) method [64]. According to MM/GBSA, the binding energy values between receptors and ligands can first be assigned to each residue, then be subdivided into internal energy in vacuum (including polar electrostatic interactions and non-polar van der Waals interactions), polar solvation energy and non-polar solvation energy, and eventually be decomposed into the main chain and side chain of each residue. Here, the polar and non-polar solvation energy can be calculated with generalized Born (GB) model and LCPO algorithm [65], respectively.
The identification of hot spots between the PPIs plays a key part in the detection of active sites of receptors. By now, hierarchical clustering (HC) analysis has been an efficient technology for determining the hot spots and common binding characteristics [66]. In this work, key residues were identified by HC analysis based on energy contributions per amino acid using R statistical package. Manhattan distance is used to describe the similarity levels among vectors. It is defined as:
where i indicates the dimensional index of individual binding energies a and b at residual level. To implement cluster discrimination, the Ward’s minimum variance method was adopted here [67]. Finally, the calculation result files were processed using the online tree generator iTOL to formulate the hierarchical tree graph shown in color-coded modes [68]. Through energy decomposition and HC analyses, the key residues in the PPIs of S protein with three essential partners can be obtained.
Solvent accessible surface area (SASA) calculation
SASA is one of the key parameters to be considered in the study of biomolecular recognition [69, 70]. SASA is generally defined as the surface area formed by a water probe ball rolling around the surface of biomolecules. The larger the SASA, the weaker the hydrophobic effect. In the actual calculation, SASA can be assigned into protein residual level. It is worth mentioning that some amino acids are buried in the proteins deeply and the corresponding SASA is zero. In addition, the buried SASA (Sburied) is an important criterion for evaluating the strength and effectiveness of molecular recognition between proteins. The computational formula is as follows:
Here, SA and SC are used to describe the SASAs of spike protein and complexes, respectively. In this work, the SASA of four trimers (i.e., WT, Delta, Mu, Omicron) and their complexes with hACE2/TMPRSS2/C121 can be computed from MD trajectories with gmx sasa module using the algorithms of Eisenhaber et al. [71].
Self-defined parameters
In order to more accurately describe the mutual recognition between S protein and three essential partners (i.e., hACE2, TMPRSS2, and C121), the following three self-defined parameters are provided. (1) Water basin: formed by the part near mass center in the up-state RBD (chain A, E516-G566) and the adjacent HR1 (chain C, G910-D985) and NTD (chain B, N30-F55), in which the amount of water varies with the open and closed state of RBD [72]. In water basin, the less the amount of water is, the easier it is for RBD to maintain the up-state and bind with hACE2, and not vice versa. (2) Tetrahedron height: the catalytic triplet of TMPRSS2 (i.e., H296, S441, S460) were found to interact with the cleavage sites (i.e., R815, S816) of SARS-CoV-2 spike [73]. In the spike complexes with TMPRSS2, tetrahedron height representing vertical distance from the center points of R815/S816 to the plane formed by Cα atoms of the catalytic triplet. The size and stability of tetrahedron height can reflect the affinity of S protein and TMPRSS2 to a certain extent: the farther the distance, the harder it is to bind. (3) Approaching angle: the normal vectors of the three planes (i.e., S2 region horizontal plane, antibody plane interacting with RBD, and RBD plane interacting with antibodies) were defined, and approaching angle 1 is formed by the first two vectors, with approaching angle 2 by the latter vectors. The Cα of three amino acids is connected to form a plane, and we define three such planes. The horizontal plane of S2 region is defined by T998 of the three chains (located in the stable spiral region). The other two are represented respectively by three points of the interface between RBD and C121 (RBD: G446, E484, V503; C121: T28, V54, S88). Different types of neutralizing antibodies have special approaching angle to S protein interface, which is related to its neutralization effect [74].
Results
Introduction to simulation systems
The interactions of WT S protein and three mutants respectively with three partners (i.e., hACE2, TMPRSS2, and C121) was explored in this work. Figure 2 shows their molecular structure and the list of mutation sites. Spike protein is composed of three chains with residue range from A27 to S1147. According to the order of residues, the noteworthy domains in spike are as follows: (1) RBD (R319-N540), its transition from down- to up-conformation is a key step before binding with hACE2, being a hot spot targeted by mainstream antibodies; (2) NTD (S13-F318), the target region of another potent antibody, although its specific biological function is unclear; (3) 630 loop (V620-S640), the transition between ordered and disordered conformation affects the opening and closing efficiency of RBD; (4) S2’ (R815/S816), as the cleavage site of TMPRSS2, it is closely related to virus replication; (5) FPPR (F823-P862), it not only affects the recognition by TMPRSS2, but also acts synergistically with 630 loop to affect the transition of down- to up-conformation of RBD; (6) HR1 (G910-D985), as the most important part of the transmembrane helices, they insert the fusion peptides into the target cell membrane, by undergoing significant conformational rearrangement. In terms of mutation sites, compared with WT, Delta possesses 7 mutations and 2 deletions, with Mu having 9 mutations, as well as Omicron having 30 mutations, 5 deletions, and 1 insertion. Excessive mutations suggest that Omicron spike may exhibit novel characteristics including substrate recognition, conformational change, and immunogenicity.
Molecular structure of S protein trimer and its essential partners (A), as well as the list of mutation sites (B). The circled part is the binding site for the three essential partners, where the red parts of TMPRSS2, C121, and hACE2 represent the recognition interface by spike. S protein trimer is shown using surface model, and solid-ribbon model is added to the outermost chain A. The binding regions for the three partners with spike are Q24-L45/L79-Y83/T324-D355 of hACE2, H296-A486 of TMPRSS2, and H: G26-T30/W50-S75/Y95-Y112 of C121, which are represented in red in the upper right a/b/c panels
Overall structural convergence
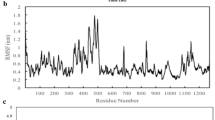
Figure 3A, S1A, and S2A show the potential energy of all 12 systems over simulation time, as well as the mean and standard deviation. The systems all reach potential energy equilibrium after 5 ns, indicating that the simulation process is smooth and the trajectory is reliable. By measuring the overall difference between the conformational snapshot and the first frame, RMSD values can be used to effectively evaluate whether the trajectories of biomolecules have reached the equilibrium state in the MD simulation. As shown from Fig. 3B, S1B, and S2B, RMSD values keep relatively stable after 50 ns, and the equilibrium trajectory in this period can be used for subsequent analyses of conformational change and molecular recognition. Overall, the fluctuation of RMSD values is similar for the 12 investigated systems, ranging from 3.47 Å (Mu_TMPRSS2) to 7.41 Å (Mu_hACE2). The relatively high RMSD value is due to the large simulation system with more than 3500 residues and excessive number of mutations.
Conformation convergence parameters for MD simulations of the WT_hACE2, Delta_hACE2, Mu_hACE2, and Omicron_hACE2 systems. The variation of potential energy (A) and RMSD (B) with time; RMSF distribution at residue level of S protein chain A in the four systems (C); RMSF correlation of S protein chain A between Delta_hACE2 and WT_hACE2 (D)
RMSF can provide flexibility difference of the simulation system at residual level. As shown in Fig. 3C, the RMSF distribution in four Spike_hACE2 systems are highly similar. Specially, HR1(G910-D985) has the lowest flexibility, which is closely related to the high rigidity of transmembrane helical [75]. Moreover, hACE2 receptor binding motif (RBM, N437-P507) in S protein also shows low flexibility, which is consistent with Shang et al.’s work that the greater rigidity in RBM is conducive to hACE2 binding [76]. In the Spike_TMPRSS2 system, the two regions above still maintain high conservatism and low flexibility. The flexibility of Delta and Mu mutants at S2’ (R815/S816) was significantly lower than WT, which may affect the affinity and cutting ability of TMPRSS2 to spike [73]. There are some differences in NTD (S13-F318) flexibility among the Spike_TMPRSS2 systems, as well as among the four Spike_C121 systems (see Fig. S2). It should also be mentioned that the flexibility differences in the NTD region may determine the effectiveness of NTD-targeted antibodies [77]. In addition, the good correlation between RMSF values of Cα atoms in different S protein systems (see Fig. 2D, S1D, and S2D) further proves the reliability of the twelve MD simulation trajectories.
Molecular recognition of S protein and hACE2
hACE2 is a carboxypeptidase with 805 residues containing an N-terminal peptidase domain and a C-terminal collectrin-like domain [78]. As a component of the renin–angiotensin–aldosterone system, hACE2 is primarily responsible for maintaining angiotensin levels [16, 79]. Previous studies have shown that hACE2 is the receptor protein for SARS-COV-2 to invade cells, and their binding is a key step in triggering COVID-19 infection [16, 17]. The recognition interface between S protein RBD and hACE2 is mainly composed of the concave surface of RBM region and the N-terminal helix of the receptor [80]. Compared with SARS-CoV, several amino acid substitutions were observed in SARS-COV-2 RBD, suggesting that mutations in spike could alter viral tropism, including introduction of new hosts or increased transmission of the virus [81]. Subsequently, according to Ortega et al.’s work, these changes are mainly related to enhanced hACE2 interactions [8].
The up-state of RBD is the prerequisite for the stable recognition of S protein and hACE2. To investigate whether the three mutations affect the conformational state of RBD, the amount of water in the water basin was monitored over time. As shown from Fig. 4B, the amount of water in S protein mutants is much less than WT, especially Delta. For similar systems, solvent effect is not conducive to the maintenance of pocket structure, for example, the inner pocket of the protein is more hydrophobic [86]. Obviously, the ability of RBD in the four spike systems maintaining up-state is Delta > Omicron ≈ Mu > WT. To further verify the above viewpoints, conformation cluster was performed based on MD trajectories of four S protein trimers, and the representative conformation with the highest proportion (WT 74%, Delta 68%, Mu 66%, Omicron 72%) was selected for superposition with the initial structure of WT. As shown in Fig. 4C, compared with the initial structure, the RBD of mutants has a smaller migration displacement than WT, better maintaining the original up-state and the potential of hACE2 recognition, thus enhancing virus transmission.
Previous studies have shown that the state of RBD is not only affected by water, but also related to the conformation of 630-loop (V620 to S640) [82]. No matter whether RBD is in up- or down-state, the 630-loop has internal ordered-disordered dynamic switch to a certain extent. The ordered 630-loop helps RBD sustain the down-state [83], while the disordered 630-loop is more likely to stay in the up-conformation, which is conducive to the recognition by hACE2. The MD trajectories of four S protein trimers were clustered, and the representative conformations (see Fig. 4C) were obtained. Subsequently, the comparison of 630-loops showed that the mutants (i.e., Delta, Mu, and Omicron) were more disordered than WT (see Fig. S3). It is consistent with the conclusion that mutants exhibit higher infectivity and richer conformation after fusion with human host cell [84].
Fig. S4 shows the top ten residues with the largest buried area (Sburied) in the WT_hACE2, Delta_hACE2, Mu_hACE2, and Omicron_hACE2 systems. The Sburied can effectively represent the intensity of receptor-ligand interactions, and the higher the value, the stronger the affinity. Amino acids occurring over two times (i.e., K417, L455, F486, Y489, Q/R483, Q/R498, T500, N/Y501, and Y/H505) are defined as high-frequency contact residues, and vice versa are referred to as low-frequency contact ones (i.e., Y449, F456, A475, N487, S494, G/S496, and G502) [85]. Compared with WT_hACE2, the contact residues in Mu_hACE2 were significantly changed (low-frequency contact residues A475, N487, S494 and mutation site Y501 were introduced), but the two systems still maintained the similar Sburied (~ 5.7 Å2). Five mutated residues (i.e., R493, S496, R498, Y501, and H505) appeared in the hACE2 recognition region of Omicron spike, resulting in a larger Sburied (~ 6.87 Å2) than WT (~ 5.74 Å2) and thus a greater affinity [86]. In addition, the Sburied (~ 6.24 Å2) of Delta_hACE2 is slightly larger than that of WT_hACE2 (~ 5.74 Å2), which may be related to the occurrence of low-frequency contact residues Y449 and G496. It can be inferred from the Sburied results that Omicron and Delta should have better affinity with hACE2 than the WT and Mu systems.
The parameter of binding free energy can accurately characterize the binding ability between molecules. Figure 5 shows the binding free energy of four Spike_hACE2 complexes. Mutants, especially Delta and Omicron, have a significantly higher affinity to hACE2 than WT, which is consistent with the Sburied analyses above. The common feature of receptor-ligand recognition can be found through HC analysis on the energy decomposition values of key residues. As shown from Fig. 5B, four different clusters (i.e., A1–A4) were obtained from energy contributions of the key residues. In WT S protein, cluster A4 (Q493, Q498, N501, Y505) provided the most favorable energy contribution to molecular recognition by hACE2, descending sequentially to A3 (L455, F486, Y489, T500), A1 (K417, Y449, S496), and A2 (K444, R454, A475, G502). Interestingly, the role of these residues in A4 was somewhat diminished in the Delta, Mu, and Omicron mutants, but was effectively compensated by other clusters. Specifically, Delta reflects the importance of cluster A1, Mu improves affinity by increasing the contribution of K417, Y489, Y501, etc., while Omicron mainly relies on R493 [93]. These key residues extracted from HC analysis will contribute to the subsequent design and development of therapeutic antibodies selectively targeting different S protein mutants.
The binding modes (see Fig. S5) complement the details of key residue interactions, precisely duplicating and enriching the results of HC analysis (see Fig. 5). Although hACE2 is stably recognized by different residues, K353 appears in different spike mutants showing a certain conservatism [73]. It is worth mentioning that the N501Y mutation enhances the interaction with K353 by introducing pi-alkyl, which has been confirmed by previous studies [87]. As WT mutated into Omicron, the intermolecular interactions of S protein gradually diversified from a single hydrogen bond. Although both electrostatic and hydrophobic interactions drive the binding of S protein to hACE2 [88], the former including hydrogen bonds is relatively dominant [89]. In the Omicron system, RBD mutation forms a larger positively charged region, which effectively binds to the negatively charged residues of hACE2 to increase their affinity; there are strong salt bridges of R493 with D38 and E35; Y505H weakens the affinity with hACE2, but Q493R greatly reverses this disadvantage [405].
Molecular recognition of S protein and TMPRSS2
SARS-CoV-2 enters the host cell in two ways, TMPRSS2-mediated plasma membrane fusion and the endosomal pathway [90]. Human TMPRSS2 consists of 492 residues, and its C-terminal peptidase S1 domain (I256-Q487) possesses the typical structural characteristics of chymotrypsin family serine proteases, showing the core cleavage function [91, 92]. Substrate binding sites (D435, S460, G462) and catalytic active sites (H296, D345, S441) both are considered to be key residues of TMPRSS2, and the latter is closely related to its cleavage performance. In the presence and mediation of TMPRSS2 and other enzymes, S1/S2 and S2’ sites both are cleaved to expose various fragments, initiating the plasma membrane fusion path [93]. Compared with SARS-CoV, the new emergence of multi-alkali cleavage site (PBCS) in SARS-CoV-2 makes its transmission and pathogenicity more dependent on TMPRSS2 [94,95,96,97]. Therefore, study on molecular recognition of TMPRSS2 and S protein will contribute to a deep understanding of the enzymatic cleavage process at the atomic level, which will guide the design and development of inhibitors targeting TMPRSS2.
So far, the full length of TMPRSS2 and its complex with S protein both have not been resolved, which may be related to the high percentage of helicity and surface hydrophobicity [98]. The S protein cleavage site R815/S816 was defined as the active residues of SARS-CoV-2, and the catalytic active site (H196, D345, S441) and the substrate binding site (D435, S460, G462) were identified as the active residues of TMPRSS2. In 2020, fortunately, Hussain et al.’s study revealed a reasonable complex model for Spike_TMPRSS2 via the HADDOCK online server [73]. We used TMPRSS2 in the complex of partial length human TMPRSS2 and inhibitor nafamostat 9 (PDB ID: 7MEQ) as a model [56]. Using the same key residues mentioned above to construct the complex structure with HADDOCK2.4 web server. The scoring of HADDOCK is a linear combination function of van der Waals energy, electrostatic energy, desolvation energy, constrained failure energy, and buried surface area. The cluster with the lowest score is selected for further visual evaluation to obtain the model.
The S2 segment (i.e., V687-G1273) is a relatively low molecular flexibility region in S protein trimer with small structural changes [82]. In order to explore structural changes of S protein caused by Delta/Mu/Omicron mutations, their monomer structures were superimposed using the S2 fragment with almost unchanged conformation as a reference. As shown from Fig. 6A, the distances between S protein and TMPRSS2 in the Omicron_TMPRSS2, WT_TMPRSS2, Delta_TMPRSS2, and Mu_TMPRSS2 systems decrease successively; from another perspective, TMPRSS2 in Mu_TMPRSS2 and Delta_TMPRSS2 also rotated, gradually approaching the chain B of S protein. The D796-C851 segment of S protein is involved in the recognition by TMPRSS2, where region 1 (i.e., C840-C851) in the proximal position of the fusion peptide (FP) undergoes significant conformational changes, while region 2 (D796-V826) maintains relatively stable (see Fig. 6B). By contrast, region 1 and TMPRSS2 both show almost the same motion direction, indicating that region 1 dominates or at least participates in TMPRSS2’s motion pattern.
Conformational changes of Spike_TMPRSS2 binding interface. A The S protein monomer structures of the WT_TMPRSS2, Delta_TMPRSS2, Mu_TMPRSS2, and Omicron_TMPRSS2 systems were superimposed using the S2 fragment with almost unchanged conformation as a reference; B the binding site of spike with TMPRSS2 is divided into two regions that are presented in a/b panels
Fig. S6 shows the top five residues with the largest buried area (Sburied) in WT_/Delta_/Mu_/Omicron_TMPRSS2, of which S810 is present in all systems. The Sburied in the WT_/Delta_/Mu_TMPRSS2 systems increased successively, with average values of 2.59, 3.39, and 3.47 Å2, respectively, which is consistent with the decreasing distance between spike and TMPRSS2. In the Omicron_TMPRSS2 system, the above distance is the farthest, surprisingly accompanied by the largest Sburied. It suggests that the Omicron mutants containing more than 30 mutations exhibit different recognition patterns with TMPRSS2 from those of WT, Delta, and Mu. Compared with WT_TMPRSS2, Omicron_TMPRSS2 shows the most significant changes in key residues, including low-frequency contact residues I794, Y796 (mutation site), and P809. In general, the Sburied value between TMPRSS2 and mutants (i.e., Delta, Mu, and Omicron) was similar and all greater than that of WT_TMPRSS2, which was consistent with the mutants showing more significant cleavage and immune escape.
According to the prediction of binding free energy of S protein trimer to TMPRSS2, the three mutants have higher affinity than the WT. Detailed interaction details can be obtained by HC analysis of binding energies from key residues. As shown from Fig. S7B, the key residues affecting the recognition of spike by TMPRSS2 can be divided into four clusters (i.e., B1–B4) from the perspective of binding energy. In terms of WT_TMPRSS2, the B4 cluster (i.e., P793, P812, L821, F833) is most conducive to molecular recognition by TMPRSS2, but its role in three mutants is weakened. In the Delta_TMPRSS2 and Mu_TMPRSS2 systems, the clusters B1 (such as B-N657 and B-V656) and B3 (such as B-N679 and B-H655) belonging to S protein chain B both contribute to enhance the binding ability of TMPRSS2. It is worth mentioning that most residues in clusters B1 and B2 are located near the S1/S2 cleavage sites. Presumably, TMPRSS2 cannot only hydrolyze specific S2’ sites, but also further split S1/S2 sites through translocation mechanism, which improves the efficiency of being cut for the Mu and Delta mutants. In Omicron_TMPRSS2, the D796Y mutation enhanced intermolecular affinity, which was related to the newly introduced stronger hydrophobic interaction between TMPRSS2-Y416 and Spike-Y796 (see Fig. S8). It can also be seen from Fig. S8 that hydrogen bonding and hydrophobic interaction both are the main driving forces for the formation of Spike_TMPRSS2 complex.
The transmembrane efficiency and mode of SARS-CoV-2 are not only dependent on the binding ability of TMPRSS2 to S protein, but also closely related to its cleavage ability [99]. The principle of TMPRSS2 enzymatic reaction is that the catalytic triplet (H296, D345, and S441) dominates the cleavage of spike S2’ site (R815/S816) and exposes a series of hydrophobic residues, which contributes to viral fusion and entry into host cells [73]. Obviously, in the tetrahedron composed of H296, D345, S441, and R815/S816 centroids as vertices, the self-defined tetrahedron height can effectively characterize the catalytic efficiency of TMPRSS2: the smaller the distance and fluctuation amplitude, the stronger the cutting potency. As shown from Fig. 7B, the average values of tetrahedron height in the WT_/Delta_/Mu_/Omicron_TMPRSS2 systems are 5.68/7.13/13.43/14.51 Å, respectively. Specifically, Delta_TMPRSS2 and WT_TMPRSS2 both possess a short distance between S2’ site and catalytic triplet with low fluctuation, showing a potentially strong cleavage ability of TMPRSS2. Given that the distance of triplet-S’ site keeps large in the early stage of MD simulation while dropping sharply after 80 ns, the cutting efficiency of TMPRSS2 against Mu S protein is relatively weak. For Omicron_TMPRSS2, the corresponding distance and fluctuation both are the largest, which is not conducive for spike to be cleaved into FP and then integrated into host cells. In a word, the catalytic triplet analysis shows that the cleavage potency of TMPRSS2 on S protein from high to low is WT > Delta > Mu > Omicron.
A comprehensive consideration of the binding energy and cleavage efficiency of TMPRSS2 and S protein is crucial for the correct evaluation of the infectivity and virulence from different SARS-CoV-2 virus strains. Specially, WT is easily cut by TMPRSS2, but its low binding ability greatly reduces infectivity and toxicity. The Delta mutants showed strong viral replication and pathogenicity, suggesting that TMPRSS2 inhibitors may have high antiviral potential against this type of strain [88]. Although Mu spike has the strongest binding capacity to TMPRSS2, while accompanied by a weak cutting ability, so the transmembrane efficiency is low, which may be one of the reasons for the narrow epidemic area of Mu virus strain. Omicron obviously abandoned the transmembrane of TMPRSS2 pathway with much lower binding ability and cutting ability than that of Delta, which was consistent with the significant decrease of syncytial formation [100]. In other words, the endosomal pathway has emerged as a novel option for transmembrane of Omicron strain [101]. Mutations of more than 30 residues resulted in reduced S1/S2 cleavage efficiency, accompanied by an increase in the positive charge of the S protein, which all contribute to Omicron mutants favoring endosomal pathway [102].
Molecular recognition of S protein and C121
The purpose of vaccination for COVID-19 patients is to make the body produce corresponding antibodies, which is the main strategy against SARS-CoV-2. Study on escape the mechanism of viral mutants to antibodies can provide a basis for the development of vaccine and the selection of clinical diagnosis and treatment. C121, encoded by VH3-53, is a strongly neutralizing antibody that can be repeatedly found in the plasma of most convalescent or vaccinated patients [103]. Current neutralization mechanisms of antibodies targeting SARS-CoV-2 are diverse, such as blocking receptor binding, steric hindrance, locking S protein in down-conformation, and so on. Antibody C121 binds to RBD’s receptor-binding ridge (i.e., the main epitopes) and overlaps with the polar and hydrophobic residues in the plane responsible for the interaction with hACE2, exerting inhibitory activity through the mechanism of competitive binding [57]. Currently, the most mainstream antibodies used to treat COVID-19 are RBD-directed type. In addition to antibody C121, there are some antibodies with the same epitope, such as C002, C009, 5A6, and COVA2-39 [63].
Antibodies targeting different epitopes bind to S protein at different approaching angles, whereas those from the same epitope having similar approaching angles [67]. Given the right approaching angle is the key factor for antibody C121 to capture spike antigen epitope, Fig. 8 comparatively shows their variation over simulation time in the WT_C121, Delta_C121, Mu_C121, and Omicron_C121 systems. In terms of approaching angle 1 used to characterize the recognition of spike by C121 at the macro level, the values from high to low are Omicron_C121 > Mu_C121 > Delta_C121 ≈ WT_C121; while for the approaching angle 2 more suitable for representing the C121-RBD recognition at the micro level, the systems are Omicron_C121 > WT_C121 ≈ Delta_C121 ≈ Mu in descending order. By comparing the deviation of approaching angles in three mutants to those in the WT, Omicron mutants shows obvious immune escape from this kind of RBD-directed antibodies.
Fig. S10 shows the top 10 residues with the largest contribution of Sburied on the recognition interface in the four Spike_C121 systems. The mean Sburied values of WT_C121, Delta_C121, Mu_C121, and Omicron_C121 are 6.51, 6.12, 5.3, and 5.22 Å2, initially showing the binding potency of descending order. Key residues with high frequency including Y449, L455, F456, E484, F486, Y489, F490, Q493, and Q498 have all been reported previously [63]. The key amino acids in WT_C121 and Delta_C121 are almost the same, and all of them appear at high frequencies except T500, which also guarantees their similar Sburied values. The key residues in the Omicron_C121 system change significantly. some high frequency residues (e.g., L455 and F456) that appear in the other three systems are no longer present, while new contribution sites (e.g., G485, S494, and C488) and mutation residues (e.g., E484A and Q498R) both lead to the minimal Sburied. The variation degree of key residues in Mu is between Delta and Omicron, with P479 only appearing in this system.
Several studies have confirmed that residue mutations (e.g., K417N, E484K, L452R, N501Y) located at the interface of RBD (R319-N540) can reduce the neutralizing activity of antibodies to varying degrees [63, 104, 105]. By superimposing the RBMs (N437-P507) of the four Spike_C121 complexes, it is found that there is significant conformational change in the I472-F490 loop, which also covers some key epitope residues (e.g., E484, F486, Y489, and F490). In fact, a series of previous studies have also shown that the I472-F490 loop exhibits great flexibility in the Alpha, Beta, Delta, Mu, and Omicron mutants. In order to quantitatively compare the conformational changes of the loop regions, the changes in the distance between Ca atoms of residues P479, E484, and Q493 were measured for the four Spike_C121 systems (see Fig. 9). These residues were selected for the following reasons: P479 is a unique contact residue in the Mu system; Q493, not far from the loop, is a good reference point with a constant position; the high-frequency contact residue E/K/A484 appears in all spike systems except Mu. The mean E484-Q493 distances of Mu_C121 and WT_121 are 22.41 and 20.60 Å, while they decrease to 14.22 and 14.32 Å for Delta_C121 and Omicron_C121. In addition, Mu_C121 has a shorter P479-Q493 distance than the other three systems with a decrease of about 3 Å, which is consistent with the fact that P479 is only the key contact residue of Mu_C121. In summary, the I472-F490 loop in the four Spike_C121 systems underwent obvious conformational change, which greatly affected the Sburied value of the interface, intermolecular binding force, and antibody efficacy [106].
Fig. S11 shows binding free energies of spike trimer to antibody C121. Under the same calculation parameters, the descending order of the predicted binding capacity with C121 was WT, Delta, Mu, and Omicron. Based on energy decomposition data of key residues, three different clusters (C1–C3) especially C1 (Y489, F490, F486) and C3 (V483, E484, F456, L455, Y449) conducive to the association of S protein and C121 were provided by HC analysis. In the Omicron and Mu systems, the decrease of binding free energy is mainly due to the weakening of energy contribution of key residues in clusters C1 and C3. Mutations such as E484K in Mu and E484A/Q493R/Q498R in Omicron were not conducive to molecular recognition of SP by antibody C121. In fact, E484K not only causes spatial collisions, but also leads to a positively charged environment at the antibody binding site, while E484A weakens some of the original hydrogen bonds of E484K [107]. According to the binding modes of Spike_C121, the hydrogen bond and hydrophobic interactions of Mu_C121 and Omicron_C121 were partially damaged compared with WT_C121, which reasonably explains the significant decrease in affinity between the viral mutants and the antibody.
Comparative molecular dynamics simulations were performed for 12 complexes. Potential energy, RMSD, and RMSF analyses all show that MD simulation trajectories are stable and reliable, and the flexibility of S protein RBM and S2’ cleavage sites decreased slightly with the binding of hACE2 and TMPRSS2. In the Spike_hACE2 system, the amount of water in the basin is WT > Mu ≈ Omicron > Delta. The order of 630 loop is WT > Mu > Delta > Omicron. The Sburied of Omicron and Delta are larger than Mu and WT. The affinity of mutants to hACE2 was higher than that of WT, and Delta was the strongest. Omicron does not rely on traditional key residues to maintain its affinity with hACE2, but R493. In the Spike_TMPRSS2 system, mutants use a different binding pattern than WT to recognize TMPRSS2. Because of the shift effect, the mutant may have the ability of double-site cutting. The short distance between S2’ site and catalytic triplet corresponding to the increase of Sburied is reflected in Mu and Delta systems. On the contrary, Omicron has a catalytic triplet with an increased Sburied but far away from TMPRSS2. The calculated results of binding free energy show that Mu > Delta > Omicron > WT. In Spike_C121 system, both Mu and Omicron have strong approaching angle fluctuation, especially Omicron. The average E484-Q493 distance of Delta_C121 and Omicron_C121 is significantly shorter, and the P479-Q493 distance of Mu_C121 is shorter than that of the other three systems, reducing by about 3 Å. The affinity of the mutants to C121 became weaker, among which Omicron was the weakest. HC analysis showed that E484, Q493, and Q498 had the most significant effect on affinity.
Discussion
SARS-CoV-2 spike protein is first recognized and adsorbed by hACE2, then cleaved by proteases such as TMPRSS2, and finally enters human host cells through virus-cell membrane fusion and endocytosis. The interactions between S protein and hACE2 receptors is a pivotal step in viral replication cycle, and its transmission efficiency depends to a large extent on this process [108, 109]. The S protein mutants enhance its affinity with hACE2 receptor by increasing hydrogen bonds, buried area, electrostatic interaction, stabilizing hot spot residues, and other mechanisms, so as to make it spread faster [76]. Several studies have confirmed that the replication rate of Omicron is slower than that of Delta, because the former is mediated by endocytosis independent of TMPRSS2 cleavage modification [84, 101]. Factually, the difference in cell entry pathway is related to clinical manifestation and severity of the disease [96]. Alex et al. estimated that Omicron-induced disease severity was about half that of Delta, with less infection of human lung cells and organs [81, 101, 110, 111]. The improvement of transmembrane efficiency will contribute to syncytial formation, directly inducing stronger viral pathogenicity [112, 113]. The syncytia formed by micron spikes were significantly attenuated in the Omicron mutant, which was consistent with the results from animal experiments [100, 102].
Mu and Omicron both showed severe resistance to a class of antibodies with the same epitope as C121, because the E484K/A mutation impaired the interactions with antibodies, as reported in previous studies [114]. Moreover, Omicron accumulated 15 mutations in RBD, which provided a solid structural foundation for its obvious immune escape. A variety of RBD-directed antibodies, including REGN10933, REGN1098, COV2-2196, LY-CoV555, C135, and S309, have been shown to significantly reduce their neutralizing activity against Omicron [115]. At present, the development of NTD-directed antibodies has met some bottlenecks, such as G142D and del143-145 both leading to immune escape of Omicron strains. The former cannot stably bind to NTD-directed neutralizing monoclonal antibodies such as S2X333 due to steric hindrance; the latter Y144del has been proven to cause a significant reduction in neutralization activity of NTD-supersite antibodies [105].
Based on MD simulation of WT/Delta/Mu/Omicron S protein in complex with hACE2/TMPRSS2/C121, Table 1 comparatively lists their conformational change and molecular affinity. Generally, the large-scale epidemic of a virus is determined by transmission efficiency, pathogenicity, immune escape, and other factors. As shown from Table 1, SARS-CoV-2 variants have a potential biological balance among transmission efficiency, pathogenicity, and immune escape, rather than showing an obvious monotonic trend. As the main epidemic strain of COVID-19 in the world, Omicron shows strong immune escape and weakened pathogenicity, which undoubtedly belongs to evolutionary direction conducive to widespread spread of the virus. There is no doubt that the protective immunity on patients infected with Omicron strains is significantly reduced by vaccination [111, 116, 117]. Fortunately, booster doses of the vaccine still provide better neutralization response, and antiviral drugs remain effective against Omicron [118]. Under the severe situation of epidemic wave caused by Omicron, in addition to physical prevention such as wearing masks and reducing crowd gathering, as well as improving vaccination rate, it is necessary to explore more conserved epitopes and develop more specific vaccines and therapeutic antibodies. Epitope prediction appeared in the eyes of vaccine developers as early as the outbreak of MERS-CoV, breaking through the stagnant research and development process and providing new vaccine candidates [119]. This technology is still in continuous application and development, and has been used in a variety of research and development of specific vaccines in recent years, such as monkeypox vaccine [120] and orthohantavirus vaccine [121]. Among them, multi-epitope vaccine is an effective way to deal with the continuous mutation of the virus, which brings good news for the future development of SARS-CoV-2 vaccine.
Conclusion
To explore recognition differences and conformational changes, revealing viral evolutionary characteristics and possible vaccine inactivation mechanism is purpose of the study. In Spike_hACE2 systems, compared with WT, the mutants still maintain high affinity with hACE2 by keeping the RBD UP, increasing the entrapment area, or using new residues to provide energy (such as R493 to bring about favorable electrostatic interactions). In the Spike_TMPRSS2 systems, the increased Sburied provides enough affinity for the mutant. The inconsistency between the binding free energy of Spike_TMPRSS2 and the distance between S2’ site and catalytic triplet indicates that the binding ability of the two is separated from the cutting ability of TMPRSS2. It is clear that Delta is significantly higher than WT in terms of TMPRSS2-dominated cleavage efficiency, while Omicron may select a novel cell fusion mechanism different from other mutants. In Spike_C121 system, the high fluctuation of approaching angles and I472-F490 loop, as well as the reduced recognition area, all explain the resistance of Mu and Omicron mutants to C121 antibody. Overall, Delta strain has high infectivity and pathogenicity with weak immune escape ability; while Omicron strain possesses strong infectivity and immune escape ability with weak pathogenicity; and Mu strain is somewhere in between. This work not only proposed the possible mechanisms of infectivity, virulence, and drug resistance of the variants, but also provided theoretical guidance for the design of a new generation of therapeutic antibodies and vaccines against COVID-19.
References
Li F (2016) Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol 3(1):237–261. https://doi.org/10.1146/annurev-virology-110615-042301
Zhong NS, Zheng BJ, Li YM et al (2003) Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People’s Republic of China, in February, 2003. Lancet 362(9393):0–1358. https://doi.org/10.1016/s0140-6736(03)14630-2
De Wit E, van Doremalen N, Falzarano D et al (2016) SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol 14(8):523–534. https://doi.org/10.1038/nrmicro.2016.81
Li F (2013) Receptor recognition and cross-species infections of SARS coronavirus. Antiviral Res 100(1):246–254. https://doi.org/10.1016/j.antiviral.2013.08.014
Zaki AM, Van Boheemen S, Bestebroer TM et al (2012) Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med 367(19):1814. https://doi.org/10.1056/NEJMoa1211721
MD Shin, Shukla S, Chung YH, et al (2020) COVID-19 vaccine development and a potential nanomaterial path forward. Nat. Nanotechnol. 15(8). https://doi.org/10.1038/s41565-020-0737-y.
Rabaan AA, Al-Ahmed SH, Haque S et al (2020) SARS-CoV-2, SARS-CoV, and MERS-CoV: a comparative overview. Infez Med 28(2):174–184
Ortega JT, Serrano ML, Pujol FH, Rangel HR (2020) Role of changes in SARS-CoV-2 spike protein in the interaction with the human ACE2 receptor: an in silico analysis. EXCLI J 19:410–417. https://doi.org/10.17179/excli2020-1167
Sahin A, Erdogan A, MutluAgaoglu P et al (2020) novel coronavirus (COVID-19) outbreak: a review of the current literature. Eurasian J Med Oncol 4(1):1–7. https://doi.org/10.14744/ejmo.2020.12220
Yan R, Zhang Y, Li Y et al (2020) Structural basis for the recognition of the SARS-CoV-2 by full-length human ACE2. Sci 367(6485):eabb2762. https://doi.org/10.1126/science.abb2762
Tortorici MA, Veesler D (2019) Structural insights into coronavirus entry. Adv Virus Res 105:93–116. https://doi.org/10.1016/bs.aivir.2019.08.002
Letko M, Marzi A, Munster V (2020) Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. NAT MICROBIOL 5:562–569. https://doi.org/10.1038/s41564-020-0688-y
Wrapp D, Wang N, Corbett KS et al (2020) Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Cold Spring Harbor Lab 367:1260–1263. https://doi.org/10.1126/science.abb2507
Walls AC, Park YJ, Tortorici MA et al (2020) Structure, function and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 181(2):281–292. https://doi.org/10.1016/j.cell.2020.02.058
Hoffmann M, Kleine-Weber H, Schroeder S et al (2020) SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181(2):271-280.e8. https://doi.org/10.1016/j.cell.2020.02.052
Wan Y, Shang J, Graham R et al (2020) Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol 94(7):e00127-e220. https://doi.org/10.1128/JVI.00127-20
Sztain TE, Ahn SH, Bogetti AT et al (2021) A glycan gate controls opening of the SARS-CoV-2 spike protein. Nat Chem 13:963–968
Zhou P, Yang XL, Wang XG et al (2020) A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579(7798):270–273. https://doi.org/10.1038/s41586-020-2951-z
Yao H, Song Y, Chen Y et al (2020) Molecular architecture of the SARS-CoV-2 virus. Cell 183(3):730–738. https://doi.org/10.1016/j.cell.2020.09.018
Belouzard S, Chu VC, Whittaker GR (2009) Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc Natl Acad Sci U S A 106(14):5871–5876. https://doi.org/10.1073/pnas.0809524106
Vankadari N (2020) Structure of furin protease binding to SARS-CoV-2 spike glycoprotein and implications for potential targets and virulence. J Phys Chem Lett 11(16):6655–6663. https://doi.org/10.1021/acs.jpclett.0c01698
Cai Y, Zhang J, Xiao T et al (2020) Distinct conformational states of SARS-CoV-2 spike protein. Sci 369(6511):eabd4251. https://doi.org/10.1126/science.abd4251
Rey FA, Lok SM (2018) Common features of enveloped viruses and implications for immunogen design for next-generation vaccines. Cell 172(6):1319–1334. https://doi.org/10.1016/j.cell.2018.02.054
Dai L, Gao GF (2021) Viral targets for vaccines against COVID-19. Nat Rev Immunol 21:73–82. https://doi.org/10.1038/s41577-020-00480-0
Bestle D, Heindl MR, Limburg H et al (2020) TMPRSS2 and furin are both essential for proteolytic activation and spread of SARS-CoV-2 in human airway epithelial cells and provide promising drug targets. BioRxiv. 3:e202000786. https://doi.org/10.1101/2020.04.15.042085
COVID-19 vaccine tracker: vaccine landscape. https://vac-lshtm.shinyapps.io/ncov_vaccine_landscape/_w_ec54614e/#tab-1609-6 (accessed June 25, 2021).
COVID-19 vaccine tracker: clinical trials database. https://vac-lshtm.shinyapps.io/ncov_vaccine_landscape/_w_ec54614e/#tab-1609-2 (accessed June 25, 2021).
Lopez Bernal J, Andrews N, Gower C et al (2021) Effectiveness of COVID-19 vaccines against the B.1.617.2 (delta) variant. N Engl J Med. 385(7):585–594. https://doi.org/10.1056/NEJMc2113090
Fontanet A, Autran B, Lina B et al (2021) SARS-CoV-2 variants and ending the COVID-19 pandemic. Lancet 397(10278):952–954. https://doi.org/10.1016/S0140-6736(21)00370-6
Our world in data: coronavirus (COVID-19) vaccinations: https://ourworldindata.org/covid-vaccinations (accessed June 25, 2021).
Juno JA, Wheatley AK (2021) Boosting immunity to COVID-19 vaccines. Nat Med 27(11):1874–1875. https://doi.org/10.1038/s41591-021-01560-x
Dekervel M, Henry N, Torreggiani M et al (2021) Humoral response to a third injection of BNT162b2 vaccine in patients on maintenance haemodialysis. Oxford University Press (OUP) 14(11):2349–2355. https://doi.org/10.1093/CKJ/SFAB152
Jones BE, Brown-Augsburger PL, Corbett KS et al (2021) The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates. Sci Transl Med 13(593):eab1906. https://doi.org/10.1126/scitranslmed.abf1906
Anon, 2020. An EUA for casirivimab and imdevimab for COVID-19. Med. Lett. Drugs Ther.62 (1614), 201–202. https://publons.com/wos-op/search/?search=An+EUA+for+Casirivimab+and+Imdevimab+for+COVID-19 (accessed July 13, 2021).
Anon, 2021. An EUA for sotrovimab for treatment of COVID-19. Med. Lett. Drugs Ther.63 (1627), 97–98. https://publons.com/wos-op/search/?search=An+EUA+for+sotrovimab+for+treatment+of+COVID-19 (accessed July 13, 2021)
Drodal S, Rosik J, Lechowicz K, et al. An update on drugs with therapeutic potential for SARS-CoV-2 (COVID-19) treatment. Drug Resist. Update. 2021(January):100794. https://doi.org/10.1016/j.drup.2021.100794.
Sebba A (2008) Tocilizumab: the first interleukin-6-receptor inhibitor. Am J Health-Syst Pharm 65(15):1413–1418. https://doi.org/10.2146/ajhp070449
Stebbing J, Krishnan V, Bono SD, Ottaviani S, & Corbellino M (2020) Mechanism of baricitinib supports artificial intelligence-predicted testing in COVID-19 patients. EMBO Mol. Med. 12 (8). https://doi.org/10.15252/emmm.202012697
Cov-lineages: VOC reports. https://cov-lineages.org/index.html#global_reports. (accessed December 9, 2021)
Han P, Li L, Liu S et al (2022) Receptor binding and complex structures of human ACE2 to spike RBD from omicron and delta SARS-CoV-2. Cell 185(4):630–640. https://doi.org/10.1016/j.cell.2022.01.001
Kat A, Tn F, Az A et al (2022) Hospital admission and emergency care attendance risk for SARS-CoV-2 delta (B.1.617.2) compared with alpha (B.1.1.7) variants of concern: a cohort study. Lancet Infect. Dis. 22(1):35–42. https://doi.org/10.1016/S1473-3099(21)00475-8
Uriu K, Kimura I, Shirakawa K et al (2021) Neutralization of the SARS-CoV-2 mu variant by convalescent and vaccine serum. N Engl J Med 385(25):2397–2399. https://doi.org/10.1056/NEJMC2114706
Xie X, Han JB, Ma G et al (2021) Emerging SARS-CoV-2 B.1.621/mu variant is prominently resistant to inactivated vaccine-elicited antibodies. Zool Res 42(6):789–791. https://doi.org/10.24272/j.issn.2095-8137.2021.343
Classification of omicron (B.1.1.529): SARS-CoV-2 variant of concern, (n.d.). https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern (accessed December 9, 2021).
Science brief: omicron (B.1.1.529) variant | CDC, (n.d.). https://www.cdc.gov/coronavirus/2019-ncov/science/science-briefs/scientific-brief-omicron-variant.html (accessed December 9, 2021).
Khoury DS, Cromer D, Reynaldi A et al (2021) Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat Med 27:1205–1211. https://doi.org/10.1038/s41591-021-01377-8
Cameroni E, Bowen JE, Rosen LE et al (2021) Broadly neutralizing antibodies overcome SARS-CoV-2 omicron antigenic shift. Nature 602:664–670. https://doi.org/10.1038/s41586-021-04386-2
Guo D, Duan H, Cheng Y et al (2022) Omicron-included mutation-induced changes in epitopes of SARS-CoV-2 spike protein and effectiveness assessments of current antibodies. Mol Biomed 3(1):1–18. https://doi.org/10.1186/s43556-022-00074-3
Du X, Tang H, Gao L et al (2022) Omicron adopts a different strategy from delta and other variants to adapt to host. Signal Transduct Target Ther 7(1):1–3. https://doi.org/10.1038/s41392-022-00903-5
Artese A, Svicher V, Costa G, et al (2020) Current status of antivirals and druggable targets of SARS CoV-2 and other human pathogenic coronaviruses. Drug Resist. Update. 53. https://doi.org/10.1016/j.drup.2020.100721.
Twomey JD, Luo S, Dean AQ et al (2020) COVID-19 update: the race to therapeutic development. Drug Resist Update. 53:100733. https://doi.org/10.1016/j.drup.2020.100733
Andrew W, Bertoni M, Bienert S et al (2018) SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res W1:W296–W303. https://doi.org/10.1093/nar/gky427
Xiao T, Lu J, Zhang J et al (2021) A trimeric human angiotensin-converting enzyme 2 as an anti-SARS-CoV-2 agent in vitro. Nat Struct Mol Biol 28:202–209. https://doi.org/10.1101/2020.09.18.301952
Yuan S, Chan H, Hu Z (2017) Using PyMOL as a platform for computational drug design. Wiley Interdiscip Rev Comput Mol Sci 7(2):e1298. https://doi.org/10.1002/wcms.1298
G. C P, van, Zundert J, P. G. L. M, et al (2016) The HADDOCK2.4 web server: user-friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 2016; 428:720–725. https://doi.org/10.1016/j.jmb.2015.09.014.
Fraser BJ, Beldar S, Seitova A et al (2021) Structure, activity and inhibition of human TMPRSS2, a protease implicated in SARS-CoV-2 activation. BioRxiv 1:1–19. https://doi.org/10.1101/2021.06.23.449282
Barnes CO, Jette CA, Abernathy ME et al (2020) SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature 183(4):1024–1042. https://doi.org/10.1038/s41586-020-2852-1
Wang J, Wolf RM, Caldwell JW et al (2004) Development and testing of a general amber force field. J Comput Chem 25(9):1157–1174. https://doi.org/10.1002/jcc.20035
Case, DA, Cerutti DS, Cheatham TE, Darden TA, & Kollman PA. Amber 2017, University of California, San Francisco. https://doi.org/10.13140/RG.2.2.36172.41606.
Jorgensen WL, Chandrasekhar J, Madura JD (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79(2):926–935. https://doi.org/10.1063/1.445869
Miyamoto S, Kollman PA (1992) Settle: an analytical version of the SHAKE and RATTLE algorithm for rigid water models. J Comput Chem 13(8):952–962. https://doi.org/10.1002/jcc.540130805
Humphrey Dalke A, Schulten K (1996) VMD-visual molecular dynamics. J Mol Graph 14(1):33–38. https://doi.org/10.1016/0263-7855(96)00018-5
Kottalam J, Case DA (1990) Langevin modes of macromolecules: applications to crambin and DNA hexamers. Biopolymers 29:1409–1421. https://doi.org/10.1002/bip.360291008
Kollman PA, Massova I, Reyes C et al (2000) Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Accounts Chem Res 33(12):889–897. https://doi.org/10.1021/ar000033j
Simonson T (2001) Macromolecular electrostatics: continuum models and their growing pains. Curr Opin Struct Biol 11(2):243–252. https://doi.org/10.1016/S0959-440X(00)00197-4
Chen J, Liu X, Zhang S et al (2019) Molecular mechanism with regard to the binding selectivity of inhibitors toward FABP5 and FABP7 explored by multiple short molecular dynamics simulations and free energy analyses. Phys Chem Chem Phys 22(4):2262–2275. https://doi.org/10.1039/C9CP05704H
Letunic I, Bork P (2007) Interactive tree of life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23(1):127–128. https://doi.org/10.1093/bioinformatics/btl529
Ciccarelli FD, Doerks T, von Mering C et al (2006) Toward automatic reconstruction of a highly resolved tree of life. Sci 311(5765):1283–1287. https://doi.org/10.1126/science.1123061
David T, David L et al (2011) Characterization of protein-protein interaction interfaces from a single species. Plos One 6:e21053. https://doi.org/10.1371/journal.pone.0021053
Peri C, Morra G, Colombo G (2016) Surface energetics and protein-protein interactions: analysis and mechanistic implications. Sci Rep 6(1):24035. https://doi.org/10.1038/srep24035
Eisenhaber F, Lijnzaad P, Argos P et al (1995) The double cubic lattice method: efficient approaches to numerical integration of surface area and volume and to dot surface contouring of molecular assemblies. J Comput Chem 16(3):273–284. https://doi.org/10.1002/jcc.540160303
Stecula A, Schlessinger A, Giacomini KM et al (2017) Human concentrative nucleoside transporter 3 (hCNT3, SLC28A3) forms a cyclic homotrimer. Biochem 56(27):3475. https://doi.org/10.1021/acs.biochem.7b00339
Hussain M, Jabeen N, Amanullah A et al (2020) Molecular docking between human TMPRSS2 and SARS-CoV-2 spike protein: conformation and intermolecular interactions. AIMS Microbiol 6(3):350–360. https://doi.org/10.3934/microbiol.2020021
Cerutti G, Guo Y, Zhou T et al (2021) Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single supersite. Cell Host Microbe 29(5):819–833. https://doi.org/10.1101/2021.01.10.426120
Cai Y, Zhang J, Xiao T, Peng H, Chen B (2020) Distinct conformational states of SARS-CoV-2 spike protein. Sci 369(6511):1586–1592. https://doi.org/10.1126/science.abd4251
Shang J, Ye G, Shi K et al (2020) Structural basis of receptor recognition by SARS-CoV-2. Nature 581(7807):221–224. https://doi.org/10.1038/s41586-020-2179-y
Yu W, Wu X, Ren J et al (2020) Mechanistic insights to the binding of antibody CR3022 against RBD from SARS-CoV and HCoV-19/SARS-CoV-2: a computational study. Comb Chem High Throughput Screen 24(7):1069–1082. https://doi.org/10.2174/1386207323666201026160500
Donoghue M, Hsieh F, Baronas E et al (2000) A novel angiotensin-converting enzyme-related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1–9. Circ Res 87(5):E1-9. https://doi.org/10.2174/1386207323666201026160500
Crackower M, Sarao R, Oudit G et al (2002) Angiotensin-converting enzyme 2 is an essential regulator of heart function. Nature 417:822–828. https://doi.org/10.1038/nature00786
Lan J, Ge J, Yu J et al (2020) Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581:215–220. https://doi.org/10.1038/s41586-020-2180-5
Shang J, Wan Y, Liu C et al (2020) Structure of mouse coronavirus spike protein complexed with receptor reveals mechanism for viral entry. PLoS Pathog 16(3):e1008392. https://doi.org/10.1371/journal.ppat.1008392
Zhang J, Cai Y, Xiao T et al (2021) Structural impact on SARS-CoV-2 spike protein by D614G substitution. Sci 372(6541):525–530. https://doi.org/10.1126/science.abf2303
Jackson CB, Farzan M, Chen B et al (2022) Mechanisms of SARS-CoV-2 entry into cells. Nat Rev Mol Cell Biol 23(1):3–20. https://doi.org/10.1073/pnas.2003138117
Zhao H, Lu L, Zheng P et al (2022) SARS-CoV-2 omicron variant shows less efficient replication and fusion activity when compared with delta variant in TMPRSS2-expressed cells. Emerg Microbes Infect 11:277–283. https://doi.org/10.1080/22221751.2021.2023329
Jawad B, Adhikari P, Podgornik R, Ching WY (2021) Key interacting residues between RBD of SARS-CoV-2 and ACE2 receptor: combination of molecular dynamics simulation and density functional calculation. J Chem Inf Model 61(9):4425–4441. https://doi.org/10.1021/acs.jcim.1c00560
Csl A, Yy A, Hong CB et al (2021) Mutations on RBD of SARS-CoV-2 omicron variant result in stronger binding to human ACE2 receptor. Biochem Biophys Res Commun 590:34–41. https://doi.org/10.1016/j.bbrc.2021.12.079
Starr TN, Greaney AJ, Hilton SK et al (2020) Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell 182(5):1295–1310. https://doi.org/10.1016/j.cell.2020.08.012
Norel R, Sheinerman F, Petrey D, Honig B (2008) Electrostatic contributions to protein-protein interactions: fast energetic filters for docking and their physical basis. Protein Sci 10(11):2147–2161. https://doi.org/10.1110/ps.12901
Ali F, Kasry A, Amin M (2021) The new SARS-CoV-2 strain shows a stronger binding affinity to ACE2 due to N501Y mutation. ArXiv 10:100086. https://doi.org/10.1016/j.medidd.2021.100086
Evans J P, Liu S L (2021) Role of host factors in SARS-CoV-2 entry. J Biol Chem. 297(1). https://doi.org/10.1016/j.jbc.2021.100847.
Hussain M, Jabeen N, Amanullah A, et al (2021) Structural basis of SARS-CoV-2 spike protein priming by TMPRSS2. BioRxiv. https://doi.org/10.1101/2020.04.21.052639.
Shah B, Modi P, Sagar SR (2020) In silico studies on therapeutic agents for COVID-19: drug repurposing approach. Life Sci 252:e117652. https://doi.org/10.1016/j.lfs.2020.117652
Cardinale D, Guaitoli G, Tondi D et al (2011) Protein–protein interface-binding peptides inhibit the cancer therapy target human thymidylate synthase. Proc Natl Acad Sci U S A 108(34):E542–E549. https://doi.org/10.1073/pnas.1104829108
Papa G, Mallery DL, Albecka A, Welch LG, James LC (2021) Furin cleavage of SARS-CoV-2 spike promotes but is not essential for infection and cell-cell fusion. PLoS Pathog 17:e1009246. https://doi.org/10.1371/journal.ppat.1009246
Ou T, Mou H, Zhang L et al (2021) Hydroxychloroquine-mediated inhibition of SARS-CoV-2 entry is attenuated by TMPRSS2. PLoS Pathog 17:e1009212. https://doi.org/10.1371/journal.ppat.1009212
Saito A, Irie T, Suzuki R et al (2021) Enhanced fusogenicity and pathogenicity of SARS-CoV-2 delta P681R mutation. Nature 602:300–306. https://doi.org/10.1038/s41586-021-04266-9
Peacock TP, Goldhill DH, Zhou J et al (2021) The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat Microbiol 6:899–909. https://doi.org/10.1038/s41564-021-00908-w
Paoloni-Giacobino A, Chen H, Peitsch MC et al (1997) Cloning of the TMPRSS2 gene, which encodes a novel serine protease with transmembrane, LDLRA, and SRCR domains and maps to 21q22. 3. Genomics 44:309–320. https://doi.org/10.1006/geno.1997.4845
Koch J, Uckeley Z, Doldan P, et al (2021) TMPRSS2 expression dictates the entry route used by SARS-CoV-2 infect host cells. EMBO J. 40(16). https://doi.org/10.15252/embj.2021107821.
Gupta R (2022) SARS-CoV-2 omicron spike mediated immune escape and tropism shift. Res Sq. https://doi.org/10.21203/rs.3.rs-1191837/v.
Meng B, Abdullahi A, Ferreira IATM et al (2022) Altered TMPRSS2 usage by SARS-CoV-2 omicron impacts infectivity and fusogenicity. Nature 603:706–714. https://doi.org/10.1038/s41586-022-04474-x
Shuai H, Chan FW, Hu B et al (2022) Attenuated replication and pathogenicity of SARS-CoV-2 B.1.1.529 omicron. Nature 603:693–699. https://doi.org/10.1038/s41586-022-04442-5
Robbiani DF, Gaebler C, Muecksch F et al (2020) Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature 584:437–442. https://doi.org/10.1038/s41586-020-2456-9
Isabella F, Steven K, Rawlings D et al (2021) SARS-CoV-2 B.1.617 mutations L452 and E484Q are not synergistic for antibody evasion. J Infect Dis 224(6):989–994. https://doi.org/10.1093/infdis/jiab368
Wang R, Zhang Q, Ge J et al (2021) Analysis of SARS-CoV-2 variant mutations reveals neutralization escape mechanisms and the ability to use ACE2 receptors from additional species. Immunity 54(7):1611–1621. https://doi.org/10.1016/j.immuni.2021.06.003
Baral P, Bhattarai N, Hossen L et al (2021) Mutation-induced changes in the receptor-binding interface of the SARS-CoV-2 delta variant B.1.617.2 and implications for immune evasion. Biochem Biophys Res Commun 574:14–19. https://doi.org/10.1016/j.bbrc.2021.08.036
Wang P, Nair MS, Liu L et al (2021) Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature 593(7857):130–135. https://doi.org/10.1038/s41586-021-03398-2
Ortega JT, Serrano ML, Pujol FH et al (2020) Role of changes in SARS-CoV-2 spike protein in the interaction with the human ACE2 receptor: an in silico analysis. EXCLI J 19:410–417. https://doi.org/10.17179/excli2020-1167
Jafary F, Jafari S, Ganjalikhany MR (2021) In silico investigation of critical binding pattern in SARS-CoV-2 spike protein with angiotensin-converting enzyme 2. Sci Rep 11(1):6927. https://doi.org/10.1038/s41598-021-86380-2
Sigal A, Milo R, Jassat W (2022) Estimating disease severity of omicron and delta SARS-CoV-2 infections. Nat Rev Immunol 22(4):267–269. https://doi.org/10.1038/s41577-022-00720-5
Cong Z, Evans JP, Qu P, et al (2021) Neutralization and stability of SARS-CoV-2 omicron variant. bioRxiv. Preprint at https://doi.org/10.1101/2021.2012.2016.472934.
Braga L, Ali H, Secco I et al (2021) Drugs that inhibit TMEM16 proteins block SARS-CoV-2 spike-induced syncytia. Nature 594:88–93. https://doi.org/10.1038/s41586-021-03491-6
Edara VV, Manning KE, Ellis M, et al (2022) mRNA-1273 and BNT162b2 mRNA vaccines have reduced neutralizing activity against the SARS-CoV-2 omicron variant. Cell Rep. Med. 3(2). https://doi.org/10.1016/j.xcrm.2022.100529.
Starr TN, Greaney AJ, Addetia A et al (2021) Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Sci 371:850–854. https://doi.org/10.1126/science.abf9302
Mccallum M, Czudnochowski N, Rosen LE et al (2021) Structural basis of SARS-CoV-2 omicron immune evasion and receptor engagement. Sci 375(6583):864–868. https://doi.org/10.1126/science.abn8897
Liu L, Iketani S, Guo Y et al (2021) Striking antibody evasion manifested by the omicron variant of SARS-CoV-2. Nature 602(7898):676–681. https://doi.org/10.1038/s41586-021-04388-0
Planas D, Saunders N, Maes P et al (2021) Considerable escape of SARS-CoV-2 variant omicron to antibody neutralization. Nature 596:276–280. https://doi.org/10.1038/s41586-021-04389-z
Hu J, Peng P, Cao X et al (2022) Increased immune escape of the new SARS-CoV-2 variant of concern omicron. Cell Mol Immunol 19:293–295. https://doi.org/10.1038/s41423-021-00836-z
Shi J, Jing Z, Li S et al (2015) Epitope-based vaccine target screening against highly pathogenic MERS-CoV: an in silico approach applied to emerging infectious diseases. PLoS ONE 10(12):e0144475. https://doi.org/10.1371/journal.pone.0144475
Abdi SAH, Ali A, Sayed SF et al (2022) Multi-epitope-based vaccine candidate for monkeypox: an in silico approach. Vaccines 10:1564. https://doi.org/10.3390/vaccines10091564
Joshi A, Ray NM, Singh J et al (2022) T-cell epitope-based vaccine designing against orthohantavirus: a causative agent of deadly cardio-pulmonary disease. Netw Model Anal Health Inform Bioinforma 11(1):1–10. https://doi.org/10.1007/s13721-021-00339-x
Funding
This work was supported by the following fundings: National Natural Science Foundation of China (31870655); Sichuan Science and Technology Program (2019YFH0054); Project of Key Laboratory of Medicinal and Edible Plants Resources Development of Sichuan Education Department (10Y202104).
Author information
Authors and Affiliations
Contributions
Yan Cheng: resources, writing the original draft. Dan Zheng: conceptualization, resources. Derong Zhang: review, editing. Du Guo: visualization. Yueteng Wang: data mining. Wei Liu: project administration, supervision. Li Liang: project administration, supervision. Jianping Hu: project administration, supervision. Ting Luo: funding acquisition.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Cheng, Y., Zheng, D., Zhang, D. et al. Molecular recognition of SARS-CoV-2 spike protein with three essential partners: exploring possible immune escape mechanisms of viral mutants. J Mol Model 29, 109 (2023). https://doi.org/10.1007/s00894-023-05509-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-023-05509-4