Abstract
Context
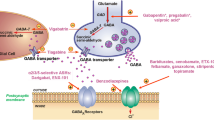
The treatment of epilepsy is associated with the inhibition of γ-aminobutyric acid-aminotransferase (GABA-AT), which suppresses the concentration of a key neurotransmitter GABA. Isosteviol, a natural bioactive molecule, has been reported to possess an anticonvulsant property. In this work, we first reported a series of C-15 fluoro isosteviol analogs which are bearing different functional groups at C-16 to investigate the interactions with GABA-AT by applying molecular docking and molecular dynamic simulation approach. The results revealed that all fluoro isosteviol analogs displayed a greater binding affinity than references vigabatrin, an FDA-approved GABA-AT inactivator, and CPP-115, which has Orphan Drug Designation status, and positioned at the same binding site as references. Furthermore, molecular dynamic (MD) simulation studies on minimum (A1), maximum (E1) binding energy score of fluoro isosteviol analogs, and isosteviol (G1) revealed their stable complex formation in terms of RMSD, RMSF, RG, and hydrogen bond formation. All analogs were found to have drug-like nature, non-toxic, >80% absorption, and the majority tend to penetrate brain-blood-barrier (BBB). The investigations found in this study can help in the development of isosteviol derivatives as drugs for the treatment of epilepsy.
Methods
The two-dimensional (2D) ligand structures were drawn using ChembioDraw Ultra 14.0. Molecular docking with Autodock4 and molecular dynamic simulation with GROMACS version 2020.1 were performed. The CHARMM27 all-atom force field was applied for writing the topology. Biovia Discovery Studio DS2021 was used for viewing and analyzing the protein-ligand complexes. The data generated from molecular dynamic simulation trajectories were plotted using the Origin® 8 software. The Open Babel software was utilized for extracting SMILEs files of all the fluoro isosteviol analogs. The drug-likeness and ADMET of the molecules were evaluated by SwissADME and ADMETlab 2.0 web tools.







Similar content being viewed by others
Availability of data and material
All data produced or analyzed during this study are incorporated in the article and supporting information. All data are available from the corresponding authors upon reasonable request.
References
Mukhopadhyay HK, Kandar CC, Das SK, et al Epilepsy and its management: a review. 7
Brigo F, Bonavita S, Leocani L et al (2020) Telemedicine and the challenge of epilepsy management at the time of COVID-19 pandemic. Epilepsy Behav 110:107164
Beghi E (2020) The epidemiology of epilepsy. Neuroepidemiology 54:185–191. https://doi.org/10.1159/000503831
高明青柳, 孝雄和田, 真知子永井 et al (1990) Increased γ-aminobutyrate aminotransferase activity in brain of patients with Alzheimer’s disease. Chem Pharm Bull (Tokyo) 38:1748–1749. https://doi.org/10.1248/cpb.38.1748
Nishino N, Fujiwara H, Noguchi-Kuno SA, Tanaka C (1988) GABAA receptor but not muscarinic receptor density was decreased in the brain of patients with Parkinson’s disease. Jpn J Pharmacol 48:331–339. https://doi.org/10.1254/jjp.48.331
Garret M, Du Z, Chazalon M et al (2018) Alteration of GABAergic neurotransmission in Huntington’s disease. CNS Neurosci Ther 24:292–300. https://doi.org/10.1111/cns.12826
Pan Y, Gerasimov MR, Kvist T et al (2012) (1 S, 3 S )-3-Amino-4-difluoromethylenyl-1-cyclopentanoic Acid ( CPP-115), a potent γ-aminobutyric acid aminotransferase inactivator for the treatment of cocaine addiction. J Med Chem 55:357–366. https://doi.org/10.1021/jm201231w
Le HV, Hawker DD, Wu R et al (2015) Design and mechanism of tetrahydrothiophene-based γ-aminobutyric acid aminotransferase inactivators. J Am Chem Soc 137:4525–4533. https://doi.org/10.1021/jacs.5b01155
Silverman RB (2018) Design and mechanism of GABA aminotransferase inactivators. Treatments for epilepsies and addictions. Chem Rev 118:4037–4070. https://doi.org/10.1021/acs.chemrev.8b00009
Cano A, Fonseca E, Ettcheto M et al (2021) Epilepsy in neurodegenerative diseases: related drugs and molecular pathways. Pharmaceuticals 14:1057. https://doi.org/10.3390/ph14101057
Storici P, Biase DD, Bossa F et al (2004) Structures of γ-aminobutyric acid (GABA) aminotransferase, a pyridoxal 5′-phosphate, and [2Fe-2S] cluster-containing enzyme, complexed with γ-ethynyl-GABA and with the antiepilepsy drug vigabatrin *. J Biol Chem 279:363–373. https://doi.org/10.1074/jbc.M305884200
Boonstra E, de Kleijn R, Colzato LS et al (2015) Neurotransmitters as food supplements: the effects of GABA on brain and behavior. Front Psychol 6:1520. https://doi.org/10.3389/fpsyg.2015.01520
Sirven JI, Noe K, Hoerth M, Drazkowski J (2012) Antiepileptic drugs 2012: recent advances and trends. Mayo Clin Proc 87:879–889. https://doi.org/10.1016/j.mayocp.2012.05.019
Cruz MP (2010) Vigabatrin (Sabril), a gamma-aminobutyric acid transaminase inhibitor for the treatment of two catastrophic seizure disorders. Pharm Ther 35:20–23
Uysal E, Konkan R, Solmaz M Psychotic episodes after vigabatrine treatment: a case report. 3
Qiu J, Silverman RB (2000) A new class of conformationally rigid analogues of 4-amino-5-halopentanoic acids, potent inactivators of γ-aminobutyric acid aminotransferase. J Med Chem 43:706–720. https://doi.org/10.1021/jm9904755
Juncosa JI, Takaya K, Le HV et al (2018) Design and mechanism of (s)-3-amino-4-(difluoromethylenyl)cyclopent-1-ene-1-carboxylic acid, a highly potent γ-aminobutyric acid aminotransferase inactivator for the treatment of addiction. J Am Chem Soc 140:2151–2164. https://doi.org/10.1021/jacs.7b10965
Clift MD, Silverman RB (2008) Synthesis and evaluation of novel aromatic substrates and competitive inhibitors of GABA aminotransferase. Bioorg Med Chem Lett 18:3122–3125. https://doi.org/10.1016/j.bmcl.2007.10.060
Quantitative structure-activity relationship and molecular docking studies of a series of quinazolinonyl analogues as inhibitors of gamma amino butyric acid aminotransferase | Elsevier Enhanced Reader. https://reader.elsevier.com/reader/sd/pii/S2090123216300790?token=2C6442EE9A6CAA78006E1C35E671719A63394A81DBD73478E0ACA3753B8C66DB9C213C6C0896BAA2A74EB3462C8D976D&originRegion=eu-west-1&originCreation=20220822144924. Accessed 22 Aug 2022
Qiu J, Stevenson SH, O’Beirn MJ, Silverman RB (1999) 2,6-Difluorophenol as a bioisostere of a carboxylic acid: bioisosteric analogues of γ-aminobutyric acid. J Med Chem 42:329–332. https://doi.org/10.1021/jm980435l
Bansal SK, Sinha BN, Khosa RL (2013) γ-Amino butyric acid analogs as novel potent GABA-AT inhibitors: molecular docking, synthesis, and biological evaluation. Med Chem Res 22:134–146. https://doi.org/10.1007/s00044-012-0023-0
Bansal SK, Sinha BN, Khosa RL (2012) Docking-based virtual screening of Schiff’s bases of GABA: a prospective to novel GABA-AT inhibitors. Med Chem Res 21:3063–3072. https://doi.org/10.1007/s00044-011-9843-6
Pinto A, Tamborini L, Pennacchietti E et al (2016) Bicyclic γ-amino acids as inhibitors of γ-aminobutyrate aminotransferase. J Enzyme Inhib Med Chem 31:295–301. https://doi.org/10.3109/14756366.2015.1021251
Tovar-Gudiño E, Guevara-Salazar JA, Bahena-Herrera JR et al (2018) Novel-substituted heterocyclic GABA analogues. Enzymatic activity against the GABA-AT enzyme from Pseudomonas fluorescens and in silico molecular modeling. Molecules 23:1128. https://doi.org/10.3390/molecules23051128
(2019) Docking studies of biologically active substances from plant extracts with anticonvulsant activity. J Appl Pharm Sci 9:66–72. https://doi.org/10.7324/JAPS.2019.90110
Asrani U, Thakur DA (2020) A comprehensive review on uses of Stevia rebaudiana plant. Clin Med 07:6
Das K (2013) Wound healing potential of aqueous crude extract of Stevia rebaudiana in mice. Rev Bras Farmacogn 23:351–357. https://doi.org/10.1590/S0102-695X2013005000011
Gupta E, Purwar S, Sundaram S, Rai GK Nutritional and therapeutic values of Stevia rebaudiana: a review. 11
Marcinek K, Krejpcio Z (2016) Stevia rebaudiana Bertoni: health promoting properties and therapeutic applications. J Für Verbraucherschutz Leb 11:3–8. https://doi.org/10.1007/s00003-015-0968-2
Sci-Hub | Computer-Aided Identification of Anticonvulsant Effect of Natural Nonnutritive Sweeteners Stevioside and Rebaudioside A. ASSAY and Drug Development Technologies, 13 (6), 313–318 | 10.1089 / adt.2015.29010.meddrrr. https://sci-hub.se/https://doi.org/10.1089/adt.2015.29010.meddrrr. Accessed 22 Aug 2022
Shah P, Westwell AD (2007) The role of fluorine in medicinal chemistry. J Enzyme Inhib Med Chem 22:527–540
Chen P, Liu G (2015) Advancements in aminofluorination of alkenes and alkynes: convenient access to β-fluoroamines. Eur J Org Chem 2015:4295–4309
Bartholomees U, Struyf T, Lauwers O et al (2016) Validation of an HPLC method for direct measurement of steviol equivalents in foods. Food Chem 190:270–275. https://doi.org/10.1016/j.foodchem.2015.05.102
Arcon JP, Modenutti CP, Avendaño D et al (2019) AutoDock bias: improving binding mode prediction and virtual screening using known protein–ligand interactions. Bioinformatics 35:3836–3838. https://doi.org/10.1093/bioinformatics/btz152
Morris GM, Huey R, Lindstrom W et al (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30:2785–2791. https://doi.org/10.1002/jcc.21256
Hsu K-C, Chen Y-F, Lin S-R, Yang J-M (2011) iGEMDOCK: a graphical environment of enhancing GEMDOCK using pharmacological interactions and post-screening analysis. BMC Bioinformatics 12:S33. https://doi.org/10.1186/1471-2105-12-S1-S33
BIOVIA DS (2017) BIOVIA Discovery Studio-BIOVIA-Dassault Systèmes®
Lee H, Le HV, Wu R et al (2015) Mechanism of inactivation of GABA aminotransferase by (E)- and (Z)-(1 S,3 S )-3-amino-4-fluoromethylenyl-1-cyclopentanoic acid. ACS Chem Biol 10:2087–2098. https://doi.org/10.1021/acschembio.5b00212
Bitencourt-Ferreira G, Pintro VO, Azevedo WF de (2019) Docking with autodock4. In: docking screens for drug discovery. Springer, pp 125–148
Parves M, Riza YM, Alam S, Jaman S (2023) Molecular dynamics-based insight of VEGFR-2 kinase domain: a combined study of pharmacophore modeling and molecular docking and dynamics. J Mol Model 29:1–15
Oshima H, Re S, Sugita Y (2020) Prediction of protein–ligand binding pose and affinity using the gREST+ FEP method. J Chem Inf Model 60:5382–5394
Saha A, Shih AY, Mirzadegan T, Seierstad M (2018) Predicting the binding of fatty acid amide hydrolase inhibitors by free energy perturbation. J Chem Theory Comput 14:5815–5822
Rizvi SMD, Shakil S, Haneef M (2013) A simple click by click protocol to perform docking: AutoDock 4.2 made easy for non-bioinformaticians. EXCLI J 12:831
Schneider G, Böhm H-J (2002) Virtual screening and fast automated docking methods. Drug Discov Today 7:64–70
Lindahl A, Hess van der S, van der Spoel D (2020) GROMACS 2020 source code. Zenodo April 302020:
Bjelkmar P, Larsson P, Cuendet MA et al (2010) Implementation of the CHARMM force field in GROMACS: analysis of protein stability effects from correction maps, virtual interaction sites, and water models. J Chem Theory Comput 6:459–466. https://doi.org/10.1021/ct900549r
Zoete V, Cuendet MA, Grosdidier A, Michielin O (2011) SwissParam: a fast force field generation tool for small organic molecules. J Comput Chem 32:2359–2368. https://doi.org/10.1002/jcc.21816
O’Boyle NM, Banck M, James CA et al (2011) Open Babel: an open chemical toolbox. J Cheminformatics 3:33. https://doi.org/10.1186/1758-2946-3-33
Daina A, Michielin O, Zoete V (2017) SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep 7:42717. https://doi.org/10.1038/srep42717
Xiong G, Wu Z, Yi J et al (2021) ADMETlab 2.0: an integrated online platform for accurate and comprehensive predictions of ADMET properties. Nucleic Acids Res 49:W5–W14. https://doi.org/10.1093/nar/gkab255
Shrivastava SK, Sinha O, Kumar M et al (2022) Synthesis, characterization, and biological evaluation of some novel ϒ-aminobutyric acid aminotransferase (GABA-AT) inhibitors. Med Chem Res 31:1594–1610. https://doi.org/10.1007/s00044-022-02935-6
Williams MA, Ladbury JE (2003) Hydrogen bonds in protein-ligand complexes. In: Böhm H-J, Schneider G (eds) Methods and principles in medicinal chemistry, 1st edn. Wiley, pp 137–161
Bojarska J, Remko M, Breza M et al (2020) A supramolecular approach to structure-based design with a focus on synthons hierarchy in ornithine-derived ligands: review, synthesis, experimental and in silico studies. Molecules 25:1135. https://doi.org/10.3390/molecules25051135
Ertl P, Rohde B, Selzer P (2000) Fast calculation of molecular polar surface area as a sum of fragment-based contributions and its application to the prediction of drug transport properties. J Med Chem 43:3714–3717. https://doi.org/10.1021/jm000942e
ImranH Khan, Patel NB, Patel VM (2018) Synthesis, in silico molecular docking and pharmacokinetic studies, in vitro antimycobacterial and antimicrobial studies of new imidozolones clubbed with thiazolidinedione. Curr Comput Aided Drug Des 14:269–283. https://doi.org/10.2174/1573409914666180516113552
Alavijeh MS, Chishty M, Qaiser MZ, Palmer AM (2005) Drug metabolism and pharmacokinetics, the blood-brain barrier, and central nervous system drug discovery. NeuroRX 2:554–571. https://doi.org/10.1602/neurorx.2.4.554
Ertl P, Schuffenhauer A (2009) Estimation of synthetic accessibility score of drug-like molecules based on molecular complexity and fragment contributions. J Cheminformatics 1:8. https://doi.org/10.1186/1758-2946-1-8
Wang S, Li Y, Xu et al (2013) Recent developments in computational prediction of hERG blockage. Curr Top Med Chem 13:1317–1326
Acknowledgements
We would like to acknowledge the National Institute of Technology Andhra Pradesh, Tadepalligudem, for providing the necessary facility to carry out the computational studies.
Author information
Authors and Affiliations
Contributions
PunamSalaria (design, conception, computational studies, data analysis, manuscript drafting, and revision), Parameswari Akshinthala (revision), Ravikumar Kapavarapu (data interpretation), and Amarendar Reddy M (design, conception, data analysis, manuscript drafting, and critical revision).
Corresponding author
Ethics declarations
Ethics approval
Not applicable.
Consent to participate
Yes.
Consent for publication
Yes.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1:
Table S1. GABA-AT (4ZSW) active siteamino acid residues involved ininteraction with ligands and bindingenergyscores. Table S2. Screening of C-15 fluoro isosteviol derivativesand standards using SwissADMEonline server to evaluate their drug-likeness, syntheticaccessibility (S.A), and oral bioavailability assays. Table S3. ADMET Screening of C-15 fluoro isosteviolderivatives using ADMETlab online designed web. Fig. S1. 2D interactionspose of isosteviol analogues: A2 (a), B1 (b), B2 (c),C1(d),C2(e),D1 (f), D2 (g),E2 (h), G2 (i), G3 (j), G4 (k) and vigabatrin (l)with 1OHW in complex form at active domain of GABA-AT enzyme. Interactions representedby different colour (m). Fig.S2. Hydrogenbond formation in,(a) A1-bound, (b) E1-bound, (c) G1-bound and(d) vigabatrin-bound complexes. Free enzyme shown in black colour. A1-GABA-AT in red, E1-GABA-AT in cyan, G1-GABA-AT in blue and vigabatrin GABA-AT in pink colour.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Salaria, P., Akshinthala, P., Kapavarapu, R. et al. Identification of novel C-15 fluoro isosteviol derivatives for GABA-AT inhibition by in silico investigations. J Mol Model 29, 76 (2023). https://doi.org/10.1007/s00894-023-05479-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-023-05479-7