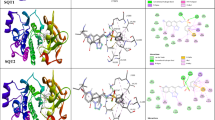
Abstract
Vascular endothelial growth factor (VEGF) and its receptor play an important role both in physiologic and pathologic angiogenesis, which is identified in ovarian cancer progression and metastasis development. The aim of the present investigation is to identify a potential vascular endothelial growth factor inhibitor which is playing a crucial role in stimulating the immunosuppressive microenvironment in tumor cells of the ovary and to examine the effectiveness of the identified inhibitor for the treatment of ovarian cancer using various in silico approaches. Twelve established VEGF inhibitors were collected from various literatures. The compound AEE788 displays great affinity towards the target protein as a result of docking study. AEE788 was further used for structure-based virtual screening in order to obtain a more structurally similar compound with high affinity. Among the 80 virtual screened compounds, CID 88265020 explicates much better affinity than the established compound AEE788. Based on molecular dynamics simulation, pharmacophore and comparative toxicity analysis of both the best established compound and the best virtual screened compound displayed a trivial variation in associated properties. The virtual screened compound CID 88265020 has a high affinity with the lowest re-rank score and holds a huge potential to inhibit the VGFR and can be implemented for prospective future investigations in ovarian cancer.














Similar content being viewed by others
Availability of data and materials
Not applicable.
Code availability
Code will be provided as per request.
References
Tew WP (2016) Ovarian cancer in the older woman. JGeriatr Oncol 7(5):354–361
Tiper IV, et al (2016) “VEGF potentiates GD3-mediated immune suppression by human ovarian cancer cells.” Clin Cancer Res: Clincanres. 2518.2015
Weiderpass E, Tyczynski JE (2015) Epidemiology of patients with ovarian cancer with and without a BRCA1/2 mutation. Mol Diag Ther 19(6):351–364
Premalata C et al (2016) Expression of VEGF-A in epithelial ovarian cancer: correlation with morphologic types, grade and clinical stage. Gulf J Oncolog 1(21):49–54
Horikawa N et al (2017) Expression of vascular endothelial growth factor in ovarian cancer inhibits tumor immunity through the accumulation of myeloid-derived suppressor cells. Clin Cancer Res 23(2):587–599
Tino AB et al (2016) Resveratrol and acetyl-resveratrol modulate activity of VEGF and IL-8 in ovarian cancer cell aggregates via attenuation of the NF-κB protein. J Ovarian Res 9(1):84
Grunewald T, Ledermann JA (2017) Targeted therapies for ovarian cancer. Best Pract Res Clin Obstet Gynaecol 41:139–152
Huang L, Huang Z, Bai Z, Xie R, Sun L, Lin K (2012) Development and strategies of VEGFR-2/KDR inhibitors. Future Med Chem 4(14):1839–1852
Traxler P, Allegrini PR, Brandt R et al (2004) AEE788: a dual family epidermal growth factor receptor/ErbB2 and vascular endothelial growth factor receptor tyrosine kinase inhibitor with antitumor and antiangiogenic activity. Cancer Res 64(14):4931–4941
Choi H-J et al (2015) Anti-vascular therapies in ovarian cancer: moving beyond anti-VEGF approaches. Cancer Metastasis Rev 34(1):19–40
Belotti D et al (2003) Matrix metalloproteinases (MMP9 and MMP2) induce the release of vascular endothelial growth factor (VEGF) by ovarian carcinoma cells: implications for ascites formation. Cancer Res 63(17):5224–5229
Wedge SR et al (2002) ZD6474 inhibits vascular endothelial growth factor signaling, angiogenesis, and tumor growth following oral administration. Cancer Res 62(16):4645–4655
Ciardiello F et al (2001) Inhibition of growth factor production and angiogenesis in human cancer cells by ZD1839 (Iressa), a selective epidermal growth factor receptor tyrosine kinase inhibitor. Clin Cancer Res 7(5):1459–1465
Traxler P et al (2004) AEE788: a dual family epidermal growth factor receptor/ErbB2 and vascular endothelial growth factor receptor tyrosine kinase inhibitor with antitumor and antiangiogenic activity. Cancer Res 64(14):4931–4941
Liu J, et al. (2018) “Assessment and management of diarrhea following VEGF receptor TKI treatment in patients with ovarian cancer.” Gynecol Oncol.
Richardson DL et al (2018) Paclitaxel with and without pazopanib for persistent or recurrent ovarian cancer: a randomized clinical trial. JAMA Oncol 4(2):196–202
Khalique S, Banerjee S (2017) Nintedanib in ovarian cancer. Expert Opin Investig Drugs 26(9):1073–1081
Orbegoso C et al (2017) The role of cediranib in ovarian cancer. Expert Opin Pharmacother 18(15):1637–1648
Zhang W et al (2017) The benefits and side effects of bevacizumab for the treatment of recurrent ovarian cancer. Curr Drug Targets 18(10):1125–1131
Azad NS et al (2008) Combination targeted therapy with sorafenib and bevacizumab results in enhanced toxicity and antitumor activity. J Clin Oncol 26(22):3709–3714
Nakamura K et al (2006) KRN951, a highly potent inhibitor of vascular endothelial growth factor receptor tyrosine kinases, has antitumor activities and affects functional vascular properties. Cancer Res 66(18):9134–9142
Xu L et al (2000) Inhibition of malignant ascites and growth of human ovarian carcinoma by oral administration of a potent inhibitor of the vascular endothelial growth factor receptor tyrosine kinases. Int J Oncol 16(3):445–499
Brozzo MS et al (2012) Thermodynamic and structural description of allosterically regulated VEGFR-2 dimerization. Blood 119(7):1781–1788
Schrodinger, LLC, NY, USA, 2009
LigPrep, Schrodinger LLC, Ney York, NY.
Prime, Schrodinger, LLC, Ney York, NY.
Protein Preparation Wizard, Schrodinger, LLC, Ney York, NY.
Qikprop, Schrodinger, LLC, Ney York, NY.
Sharma K, Patidar K, Ali MA, Patil P, Goud H, Hussain T, Nayarisseri A, Singh SK (2018) Structure-based virtual screening for the identification of high affinity compounds as potent VEGFR2 inhibitors for the treatment of renal cell carcinoma. Curr Top Med Chem 18(25):2174–2185
Sahila MM, Babitha PP, Bandaru S, Nayarisseri A, Doss VA (2015) Molecular docking based screening of GABA (A) receptor inhibitors from plant derivatives. Bioinformation 11(6):280
Vuree S, Dunna NR, Khan IA, Alharbi KK, Vishnupriya S, Soni D, Shah P, Chandok H, Yadav M, Nayarisseri A (2013) Pharmacogenomics of drug resistance in breast cancer resistance protein (BCRP) and its mutated variants. J Pharm Res 6(7):791–798
Monteiro AFM, Viana JDO, Nayarisseri A, Zondegoumba EN, Mendonça Junior FJB, Scotti MT, Scotti L (2018) Computational studies applied to flavonoids against Alzheimer’s and Parkinson’s diseases. Oxid Med Cell Longev, 2018.
Bandaru S, GangadharanSumithnath T, Sharda S, Lakhotia S, Sharma A, Jain A, Hussain T, Nayarisseri A, Kumar Singh S (2017) Helix-coil transition signatures B-Raf V600E mutation and virtual screening for inhibitors directed against mutant B-Raf. Curr Drug Metab 18(6):527–534
Kelotra A, Gokhale SM, Kelotra S, Mukadam V, Nagwanshi K, Bandaru S, Nayarisseri A, Bidwai A (2014) Alkyloxy carbonyl modified hexapeptides as a high affinity compounds for Wnt5A protein in the treatment of psoriasis. Bioinformation 10(12):743
Basak SC, Nayarisseri A, González-Díaz H, Bonchev D (2016) Editorial (Thematic Issue: chemoinformatics models for pharmaceutical design, part 1). Curr Pharm Des 22(33):5041–5042
Basak SC, Nayarisseri A, González-Díaz H, Bonchev D (2016) Editorial (Thematic Issue: Chemoinformatics models for pharmaceutical design, part 2). Curr Pharm Des 22(34):5177–5178
Prajapati L, Khandelwal R, Yogalakshmi KN, Munshi A, Nayarisseri A (2020) Computer-aided structure prediction of bluetongue virus coat protein VP2 assisted by optimized potential for liquid simulations (OPLS). Curr Top Med Chem 20(19):1720–1732
Nayarisseri A, Khandelwal R, Madhavi M, Selvaraj C, Panwar U, Sharma K, Hussain T, Singh SK (2020) Shape-based machine learning models for the potential novel COVID-19 protease inhibitors assisted by molecular dynamics simulation. Curr Top Med Chem 20(24):2146–2167
Nayarisseri A (2020) Most promising compounds for treating COVID-19 and recent trends in antimicrobial & antifungal agents. Curr Top Med Chem 20(24):2119–2125
Pochetti G, Mitro N, Lavecchia A, Gilardi F, Besker N, Scotti E, Aschi M, Re N, Fracchiolla G, Laghezza A, Tortorella P (2010) Structural insight into peroxisome proliferator-activated receptor γ binding of two ureidofibrate-like enantiomers by molecular dynamics, cofactor interaction analysis, and site-directed mutagenesis. J Med Chem 53(11):4354–4366
Bitencourt-Ferreira G, Azevedo WFD (2019) Molegro virtual docker for docking. In Docking screens for drug discovery (pp. 149–167). Humana, New York, NY.
Natchimuthu V, Bandaru S, Nayarisseri A, Ravi S (2016) Design, synthesis and computational evaluation of a novel intermediate salt of N-cyclohexyl-N-(cyclohexylcarbamoyl)-4-(trifluoromethyl) benzamide as potential potassium channel blocker in epileptic paroxysmal seizures. Comput Biol Chem 64:64–73
Bandaru S, Alvala M, Akka J, Sagurthi SR, Nayarisseri A, Kumar Singh S, Prasad Mundluru H (2016) Identification of small molecule as a high affinity β2 agonist promiscuously targeting wild and mutated (Thr164Ile) β 2 adrenergic receptor in the treatment of bronchial asthma. Curr Pharm Des 22(34):5221–5233
Majhi M, Ali MA, Limaye A, Sinha K, Bairagi P, Chouksey M, Shukla R, Kanwar N, Hussain T, Nayarisseri A, Singh SK (2018) An in silico investigation of potential EGFR inhibitors for the clinical treatment of colorectal cancer. Curr Top Med Chem 18(27):2355–2366
Khandelwal R, Chauhan AP, Bilawat S, Gandhe A, Hussain T, Hood EA, Nayarisseri A, Singh SK (2018) Structure-based virtual screening for the identification of high-affinity small molecule towards STAT3 for the clinical treatment of osteosarcoma. Curr Top Med Chem 18(29):2511–2526
Sinha K, Majhi M, Thakur G, Patidar K, Sweta J, Hussain T, Nayarisseri A, Singh SK (2018) Computer-aided drug designing for the identification of high-affinity small molecule targeting cd20 for the clinical treatment of chronic lymphocytic leukemia (CLL). Curr Top Med Chem 18(29):2527–2542
Chandrakar B, Jain A, Roy S, Gutlapalli VR, Saraf S, Suppahia A, Verma A, Tiwari A, Yadav M, Nayarisseri A (2013) Molecular modeling of acetyl-CoA carboxylase (ACC) from Jatropha curcas and virtual screening for identification of inhibitors. J Pharm Res 6(9):913–918
Nayarisseri A, Moghni SM, Yadav M, Kharate J, Sharma P, Chandok KH, Shah KP (2013) In silico investigations on HSP90 and its inhibition for the therapeutic prevention of breast cancer. J Pharm Res 7(2):150–156
Udhwani T, Mukherjee S, Sharma K, Sweta J, Khandekar N, Nayarisseri A, Singh SK (2019) Design of PD-L1 inhibitors for lung cancer. Bioinformation 15(2):139
Shukla P, Khandelwal R, Sharma D, Dhar A, Nayarisseri A, Singh SK (2019) Virtual screening of IL-6 inhibitors for idiopathic arthritis. Bioinformation 15(2):121
Nayarisseri A, Hood EA (2018) Advancement in microbial cheminformatics. Curr Top Med Chem 18(29):2459–2461
Jain D, Udhwani T, Sharma S, Gandhe A, Reddy PB, Nayarisseri A, Singh SK (2019) Design of novel JAK3 Inhibitors towards rheumatoid arthritis using molecular docking analysis. Bioinformation 15(2):68
Nayarisseri A, Singh SK (2019) Functional inhibition of VEGF and EGFR suppressors in cancer treatment. Curr Top Med Chem 19(3):178–179
Gokhale P, Chauhan APS, Arora A, Khandekar N, Nayarisseri A, Singh SK (2019) FLT3 inhibitor design using molecular docking based virtual screening for acute myeloid leukemia. Bioinformation 15(2):104
Ali MA, Vuree S, Goud H, Hussain T, Nayarisseri A, Singh SK (2019) Identification of high-affinity small molecules targeting gamma secretase for the treatment of Alzheimer’s disease. Curr Top Med Chem 19(13):1173–1187
Patidar K, Panwar U, Vuree S, Sweta J, Sandhu MK, Nayarisseri A, Singh SK (2019) An in silico approach to identify high affinity small molecule targeting m-TOR inhibitors for the clinical treatment of breast cancer. Asian Pac J Cancer Prev: APJCP 20(4):1229
Pandey N, Yadav M, Nayarisseri A, Ojha M, Prajapati J, Gupta S (2013) Cross evaluation of different classes of alpha-adrenergic receptor antagonists to identify overlapping pharmacophoric requirements. J Pharm Res 6(1):173–178
Marunnan SM, Pulikkal BP, Jabamalairaj A, Bandaru S, Yadav M, Nayarisseri A, Doss VA (2017) Development of MLR and SVM aided QSAR models to identify common SAR of GABA uptake herbal inhibitors used in the treatment of schizophrenia. Curr Neuropharmacol 15(8):1085–1092
Sweta J, Khandelwal R, Srinitha S, Pancholi R, Adhikary R, Ali MA, ... Singh SK (2019) Identification of high-affinity small molecule targeting IDH2 for the clinical treatment of acute myeloid leukemia. Asian Pac J Cancer Prev: APJCP, 20(8), 2287
Nayarisseri A (2019) Prospects of utilizing computational techniques for the treatment of human diseases. Curr Top Med Chem 19(13):1071–1074
Sharda S, Khandelwal R, Adhikary R, Sharma D, Majhi M, Hussain T, ... Singh SK (2019) A computer-aided drug designing for pharmacological inhibition of mutant ALK for the treatment of non-small cell lung cancer. Curr Top Med Chem, 19(13), 1129-1144
Limaye A, Sweta J, Madhavi M, Mudgal U, Mukherjee S, Sharma S, Hussain T, Nayarisseri A, Singh SK (2019) In silico insights on gd2: a potential target for pediatric neuroblastoma. Curr Top Med Chem 19(30):2766–2781
Nayarisseri A, Yadav M (2015) Editorial (Thematic Issue: Mechanistics in drug design-experimental molecular biology vs. molecular modeling). Curr Top Med Chem 15(1):3–4
Adhikary R, Khandelwal R, Hussain T, Nayarisseri A, Singh SK (2020) Structural insights into the molecular design of ROS1 inhibitor for the treatment of non-small cell lung cancer (NSCLC). Curr Comput Aided Drug Des 17(3):387–401
Aher A, Udhwani T, Khandelwal R, Limaye A, Hussain T, Nayarisseri A, Singh SK (2020) In silico insights on IL-6: a potential target for multicentric castleman disease. Curr Comput Aided Drug Des 16(5):641–653
Qureshi S, Khandelwal R, Madhavi M et al (2021) A multi-target drug designing for BTK, MMP9, proteasome and TAK1 for the clinical treatment of mantle cell lymphoma. Curr Top Med Chem 21(9):790–818
Yadav M, Khandelwal R, Mudgal U, Srinitha S, Khandekar N, Nayarisseri A, ... Singh SK (2019) Identification of potent VEGF inhibitors for the clinical treatment of glioblastoma, a virtual screening approach. Asian Pac J Cancer Prev: APJCP, 20(9), 2681
Nayarisseri A, Khandelwal R, Tanwar P, Madhavi M, Sharma D, Thakur G, Speck-Planche A, Singh SK (2021) Artificial intelligence, big data and machine learning approaches in precision medicine & drug discovery. Curr Drug Targets 22(6):631–655
Blessy JJ, Sharmila DJS (2015) Molecular simulation of N-acetylneuraminic acid analogs and molecular dynamics studies of cholera toxin-Neu5Gc complex. J Biomol Struct Dyn 33(5):1126–1139
Wakui N, Yoshino R, Yasuo N, Ohue M, Sekijima M (2018) Exploring the selectivity of inhibitor complexes with Bcl-2 and Bcl-XL: a molecular dynamics simulation approach. J Mol Graph Model 79:166–174
Zhang J, Scott WR, Gabel F, Wu M, Desmond R, Bae J, ... Straus SK (2017) On the quest for the elusive mechanism of action of daptomycin: binding, fusion, and oligomerization. Biochim BiophysActa (BBA)-Proteins Proteom, 1865(11), 1490–1499.
Alhadrami HA, Sayed AM, Melebari SA, Khogeer AA, Abdulaal WH, Al-Fageeh MB, ... Rateb ME (2021) Targeting allosteric sites of human aromatase: a comprehensive in-silico and in-vitro workflow to find potential plant-based anti-breast cancer therapeutics. J Enzyme Inhib Med Chem 36(1), 1334-1345
Rasul HO, Aziz BK, Ghafour DD, Kivrak A (2022) In silico molecular docking and dynamic simulation of eugenol compounds against breast cancer. J Mol Model 28(1):1–18
Cheng, Feixiong, Weihua Li, Yadi Zhou, Jie Shen, Zengrui Wu, Guixia Liu, Philip W. Lee, and Yun Tang. “admetSAR: a comprehensive source and free tool for assessment of chemical ADMET properties.” (2012): 3099–3105.
Daina A, Zoete V (2016) A boiled-egg to predict gastrointestinal absorption and brain penetration of small molecules. ChemMedChem 11(11):1117–1121
Daina A, Michielin O, Zoete V (2017) SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep 7(1):1–13
Funding
This work was supported by the Taif University Researchers Supporting Program (Project number: TURSP-2020/151), Taif University, Saudi Arabia.
The authors are grateful to the Deanship of Scientific Research, King Saud University, for funding through the Vice Deanship of Scientific Research Chairs.
SKS thank Alagappa University, Department of Biotechnology (DBT), New Delhi (No. BT/PR8138/BID/7/458/2013, dated 23rd May 2013), DST-PURSE 2nd Phase Programme Order No. SR/PURSE Phase 2/38 (G dated 21.02.2017 and FIST (SR/FST/LSI—667/2016), MHRD RUSA 1.0 and RUSA 2.0 for providing the financial assistance. UP gratefully acknowledge the Indian Council of Medical Research (ISRM/11/(19)/2017, dated 09.08.2018).
SKS thankfully acknowledges the Tamil Nadu State Council for Higher Education (TANSCHE) for the research grant (Au/S.o. (P&D): TANSCHE Projects: 117/2021).
Author information
Authors and Affiliations
Contributions
SM contributed equally to this work with MA. SM and MA were involved in molecular docking, molecular dynamics simulation, and writing — review and editing. LP, AP, AC, and MM contributed towards inhibitor collection, data curation, formal analysis, validation, and visualization. AB was involved in molecular dynamics simulation. MY, RK, AAB, and TH were involved in molecular docking, ADMET analysis, R programming analysis, and writing — review and editing. AN, AAB, TH, and SKS contributed in investigation, supervision, and writing — review and editing.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Human and animal rights
No animals/humans were used in the studies that are the basis of this research.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Sourav Mukherjee and Mohnad Abdalla have contributed equally to this work.
Rights and permissions
About this article
Cite this article
Mukherjee, S., Abdalla, M., Yadav, M. et al. Structure-Based Virtual Screening, Molecular Docking, and Molecular Dynamics Simulation of VEGF inhibitors for the clinical treatment of Ovarian Cancer. J Mol Model 28, 100 (2022). https://doi.org/10.1007/s00894-022-05081-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-022-05081-3