Abstract
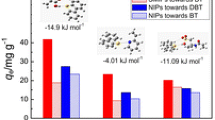
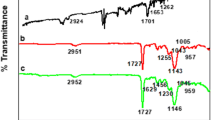
In this paper, a novel molecularly imprinted polymer (MIP) for specific adsorption of steviol glycosides was designed, and the imprinting mechanism of self-assembly system between template and monomers was clearly explored. Firstly, steviol (STE) was chosen as dummy template, and the density functional theory (DFT) at B3LYP/6–31 + G (d, p) level was used to select monomers, imprinting molar ratios, solvents, and cross-linking agents. The selectivity to five steviol glycosides was also calculated. Importantly, reduced density gradient (RDG) theory combined with atom in molecules (AIM) and infrared spectrum (IR) was applied to investigate the bonding situation and the nature of noncovalent interaction in self-assembly system. The theoretical designed results showed that the template which interacts with acrylic acid (AA) has the minimum binding energy, and the complex with the molar ratio of 1 : 4 has the most stable structure. Toluene (TL) and ethylene glycol dimethacrylate (EGDMA) were chosen as the optimal solvent and cross-linking agent, respectively. Five hydrogen bonds formed in the self-assembly system are the key forces at the adsorption sites of MIPs through the RDG and AIM analyses. The MIPs were synthesized by theoretical predictions, and the results showed that the maximum adsorption capacity towards dulcoside A is 26.17 mg/g. This work provided a theoretical direction and experimental validation for deeper researches of the MIPs for steviol glycosides. In addition, the method of RDG theory coupled with AIM and IR also could be used to analyze other imprinting formation mechanisms systematically.








Similar content being viewed by others
Data availability
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
References
Chaturvedula VS, Upreti M, Prakash I (2011) Diterpene glycosides from Stevia rebaudiana. Molecules 16(5):3552–3562. https://doi.org/10.3390/molecules16053552
Ruiz-Ruiz JC, Moguel-Ordonez YB, Segura-Campos MR (2017) Biological activity of Stevia rebaudiana Bertoni and their relationship to health. Crit Rev Food Sci Nutr 57(12):2680–2690. https://doi.org/10.1080/10408398.2015.1072083
Aranda-Gonzalez I, Moguel-Ordonez Y, Betancur-Ancona D (2015) Validation of HPLC-UV method for determination of minor glycosides contained in Stevia rebaudiana Bertoni leaves. Biomed Chromatogr 29(5):733–738. https://doi.org/10.1002/bmc.3349
Wang YH, Avula B, Tang W, Wang M, Elsohly MA, Khan IA (2015) Ultra-HPLC method for quality and adulterant assessment of steviol glycosides sweeteners - Stevia rebaudiana and stevia products. Food Addit Contam Part A Chem Anal Control Expo Risk Assess 32(5):674–685. https://doi.org/10.1080/19440049.2015.1021863
Jackson AU, Tata A, Wu C, Perry RH, Haas G, West L, Cooks RG (2009) Direct analysis of Stevia leaves for diterpene glycosides by desorption electrospray ionization mass spectrometry. Analyst 134(5):867–874. https://doi.org/10.1039/b823511b
Gardana C, Simonetti P (2018) Determination of steviol glycosides in commercial extracts of Stevia rebaudiana and sweeteners by ultra-high performance liquid chromatography Orbitrap mass spectrometry. J Chromatogr A 1578:8–14. https://doi.org/10.1016/j.chroma.2018.09.057
Wang H, Liu Y, Wei S, Yao S, Zhang J, Huang H (2016) Selective extraction and determination of fluoroquinolones in bovine milk samples with montmorillonite magnetic molecularly imprinted polymers and capillary electrophoresis. Anal Bioanal Chem 408(2):589–598. https://doi.org/10.1007/s00216-015-9140-1
Tong Z, Han Y, Gu L, Li Z, Du K, Kong G, Liu D, Peng J, Shi J (2020) Preparation and application of simetryn-imprinted nanoparticles in triazine herbicide residue analysis. J Sep Sci 43(6):1107–1118. https://doi.org/10.1002/jssc.201900739
Zhao M, Shao H, He Y, Li H, Yan M, Jiang Z, Wang J, Abd El-Aty AM, Hacimuftuoglu A, Yan F, Wang Y, She Y (2019) The determination of patulin from food samples using dual-dummy molecularly imprinted solid-phase extraction coupled with LC-MS/MS. J Chromatogr B Anal Technol Biomed Life Sci 1125:121714. https://doi.org/10.1016/j.jchromb.2019.121714
Zhu G, Cheng G, Wang P, Li W, Wang Y, Fan J (2019) Water compatible imprinted polymer prepared in water for selective solid phase extraction and determination of ciprofloxacin in real samples. Talanta 200:307–315. https://doi.org/10.1016/j.talanta.2019.03.070
Wang Q, Zhang D (2018) A novel fluorescence sensing method based on quantum dot-graphene and a molecular imprinting technique for the detection of tyramine in rice wine. Anal Methods 10(31):3884–3889. https://doi.org/10.1039/c8ay01117f
Martins RO, Gomes IC, Mendonca Telles AD, Kato L, Souza PS, Chaves AR (2020) Molecularly imprinted polymer as solid phase extraction phase for condensed tannin determination from Brazilian natural sources. J Chromatogr A 1620:460977. https://doi.org/10.1016/j.chroma.2020.460977
Sun X, Wang M, Peng J, Yang L, Wang X, Wang F, Zhang X, Wu Q, Chen R, Chen J (2019) Dummy molecularly imprinted solid phase extraction of climbazole from environmental water samples. Talanta 196:47–53. https://doi.org/10.1016/j.talanta.2018.12.017
Liu J, Wang Y, Tang S, Gao Q, Jin R (2017) Theoretical guidance for experimental research of the dicyandiamide and methacrylic acid molecular imprinted polymer. New J Chem 41(22):13370–13376. https://doi.org/10.1039/c7nj00207f
Z-q D, J-b L, S-s T, Wang Y, Li B, R-f J (2016) Theoretical design and selectivity researches on the enrofloxacin imprinted polymer. Struct Chem 27(4):1135–1142. https://doi.org/10.1007/s11224-015-0735-0
Silva CF, Borges KB, Nascimento Jr CS (2019) Computational study on acetamiprid-molecular imprinted polymer. J Mol Model 25(4):104. https://doi.org/10.1007/s00894-019-3990-y
Wang L, Yang F, Zhao X, Li Y (2020) Screening of functional monomers and solvents for the molecular imprinting of paclitaxel separation: a theoretical study. J Mol Model 26(2):26. https://doi.org/10.1007/s00894-019-4277-z
Liu JB, Wang GY, Tang SS, Gao Q, Liang DD, Jin RF (2019) Theoretical and experimental research on self-assembly system of molecularly imprinted polymers formed via chloramphenicol and methacrylic acid. J Sep Sci 42(3):769–777. https://doi.org/10.1002/jssc.201800997
Frisch MJ, Trucks GW, Schlegel HB, Scuseria MARGE, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA, Nakatsuji H, Caricato M, Li X, Hratchian HP, Izmaylov AF, Bloino J, Zheng G, Sonnenberg JL, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Vreven T, Montgomery JA, Peralta Jr. JE, Ogliaro F, Bearpark M, Heyd JJ, Brothers E, Kudin KN, Staroverov VN, Keith T, Kobayashi R, Normand J, Raghavachari K, Rendell A, Burant JC, Iyengar SS, Tomasi J, Cossi M, Rega N, Millam JM, Klene M, Knox JE, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Martin RL, Morokuma K, Zakrzewski VG, Voth GA, Salvador P, Dannenberg JJ, Dapprich S, Daniels AD, Farkas O, Foresman JB, Ortiz JV, Cioslowski J, Fox DJ (2013) Gaussian 09, Revision D.01. Gaussian, Inc., Wallingford CT,
Boys SF, Bernardi F (1970) The calculation of small molecular interactions by the differences of separate total energies. Some procedures with reduced errors. Mol Phys 19(4):553–566. https://doi.org/10.1080/00268977000101561
Zhang B, Fan X, Zhao D (2018) Computer-aided design of molecularly imprinted polymers for simultaneous detection of clenbuterol and its metabolites. Polymers (Basel) 11(1):17 https://doi.org/10.3390/polym11010017
Zhao W, Liu J, Tang S, Jin R (2020) Theoretical research of molecular imprinted polymers formed from formaldehyde and methacrylic acid. J Mol Model 26(4):88. https://doi.org/10.1007/s00894-020-04362-z
Johnson ER, Keinan S, Mori-Sánchez P, Contreras-García J, Cohen AJ, Yang W (2010) Revealing noncovalent interactions. J Am Chem Soc 132(18):6498–6506. https://doi.org/10.1021/ja100936w
Zhang X, Du X, Song J, Huang J (2019) Synthesis, crystal structure, hydrogen bonding interactions analysis of novel acyl thiourea derivative. J Phys Org Chem 33(1):e4016. https://doi.org/10.1002/poc.4016
Lu T, Chen FJ (2012) Multiwfn: a multifunctional wavefunction analyzer. J Comput Chem 33(5):580–592
Humphrey W, Dalke A, Schulten KJ (1996) VMD: visual molecular dynamics. J Mol Graph 14(1):33–38
Bader RFW, Nguyen-Dang TT, Tal Y (1981) A topological theory of molecular structure. Rep Prog Phys 44(8):893–948. https://doi.org/10.1088/0034-4885/44/8/002
Yang W, Liu L, Ni X, Zhou W, Huang W, Liu H, Xu W (2016) Computer-aided design and synthesis of magnetic molecularly imprinted polymers with high selectivity for the removal of phenol from water. J Sep Sci 39(3):503–517. https://doi.org/10.1002/jssc.201500866
Li H, Zhang W, Wu Z, Huang X, Hui A, He Y, Wang H (2020) Theoretical design, preparation, and evaluation of Ginkgolide B molecularly imprinted polymers. J Sep Sci 43(2):514–523. https://doi.org/10.1002/jssc.201900675
Huang X, Zhang W, Wu Z, Li H, Yang C, Ma W, Hui A, Zeng Q, Xiong B, Xian Z (2020) Computer simulation aided preparation of molecularly imprinted polymers for separation of bilobalide. J Mol Model 26(8):198. https://doi.org/10.1007/s00894-020-04460-y
Silva AF, Popelier PLA (2018) MP2-IQA: upscaling the analysis of topologically partitioned electron correlation. J Mol Model 24(8):201. https://doi.org/10.1007/s00894-018-3717-5
Funding
High-level talents project of Dalian City (2016RQ064); Research Foundation of Education Bureau of Liaoning Province (2016J008); National Natural Science Foundation of China (U1603285).
Author information
Authors and Affiliations
Contributions
Yajie Zhu, Yan Ding, and Yan Li designed and conceived the study; Yajie Zhu and Dongxu Tian analyzed the data and drafted the manuscript; Dongxu Tian, Yan Li, Lin-Wu Zhuang, Yinpeng Wang, and Wei Xiao provided advice and technical assistance; Jingbo Zhu provided advice and funding. Yan Ding revised the manuscript. All authors have contributed to and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Code availability
Not applicable.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOCX 13 kb)
Rights and permissions
About this article
Cite this article
Zhu, Y., Ding, Y., Tian, D. et al. Theoretical design and preparation research of molecularly imprinted polymers for steviol glycosides. J Mol Model 27, 238 (2021). https://doi.org/10.1007/s00894-021-04819-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-021-04819-9