Abstract
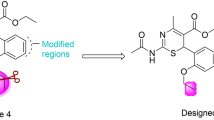
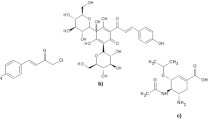
Recent outbreaks of highly pathogenic influenza strains have highlighted the need to develop new anti-influenza drugs. Here, we report an in silico study of carvone derivatives to analyze their binding modes with neuraminidase (NA) active sites. Two proposed carvone analogues, CV(A) and CV(B), with 36 designed ligands were predicted to inhibit NA (PDB ID: 3TI6) using molecular docking. The design is based on structural resemblance with the commercial inhibitor, oseltamivir (OTV), ligand polarity, and amino acid residues in the NA active sites. Docking simulations revealed that ligand A18 has the lowest energy binding (∆Gbind) value of −8.30 kcal mol-1, comparable to OTV with ∆Gbind of −8.72 kcal mol-1. A18 formed seven hydrogen bonds (H-bonds) at residues Arg292, Arg371, Asp151, Trp178, Glu227, and Tyr406, while eight H-bonds were formed by OTV with amino acids Arg118, Arg292, Arg371, Glu119, Asp151, and Arg152. Molecular dynamics (MD) simulation was conducted to compare the stability between ligand A18 and OTV with NA. Our simulation study showed that the A18-NA complex is as stable as the OTV-NA complex during the MD simulation of 50 ns through the analysis of RMSD, RMSF, total energy, hydrogen bonding, and MM/PBSA free energy calculations.








Similar content being viewed by others
References
Samson M, Pizzorno A, Abed Y, Boivin G (2013) Influenza virus resistance to neuraminidase inhibitors. Antivir Res 98(2):174–185
Wang GT, Chen Y, Wang S, Gentles R, Sowin T, Kati W, Muchmore S, Giranda V, Stewart K, Sham H, Kempf D, Laver WG (2001) Design , Synthesis , and Structural Analysis of Influenza Neuraminidase Inhibitors Containing Pyrrolidine Cores. Lett Org Chem 167:1192–1201
John P, Adabala P, Legresley EB, Bance N, Niikura M, Pinto BM (2013) Exploitation of the catalytic site and 150 cavity for Design of in fl uenza a neuraminidase inhibitors. J Org Chem 78:10867–10877
von Itzstein M (2007) The war against influenza: discovery and development of sialidase inhibitors. Nat. Rev. Drug Discov. 6:967–974
von Itzstein M, Wu W-Y, Kok GB, Pegg MS, Dyason JC, Jin B, Van Phan T, Smythe ML, White HF, Oliver SW, Colman PM, Varghese JN, Ryan DM, Woods JM, Bethell RC, Hotham VJ, Cameron JM, Penn CR (1993) Rational design of potent sialidase-based inhibitors of influenza virus replication. Nature 363:418–423
Gong J, Xu W, Zhang J (2007) Structure and functions of influenza virus neuraminidase. Curr Med Chem 14(1):113–122
Pranita SSK, Kore P, Mutha MM, Antre RV, Oswal RJ (2012) Computer-aided drug design: an innovative tool for modeling. Open J Med Chem 2(1):139–148
D’Souza C, Kanyalkar M, Joshi M, Coutinho E, Srivastava S (2009) Probing molecular level interaction of oseltamivir with H5N1-NA and model membranes by molecular docking, multinuclear NMR and DSC methods. Biochim Biophys Acta Biomembr. 1788(2):484–494
Stoll V, Stewart KD, Maring CJ, Muchmore S, Giranda V, Gui Y, Gu Y, Wang G, Chen Y, Sun M, Zhao C, Kennedy AL, Madigan DL, Xu Y, Saldivar A, Kati W, Laver G, Sowin T, Sham HL, Greer J, Kempf D (2003) Influenza neuraminidase inhibitors: structure-based design of a novel inhibitor series. Biochemistry 42(3):718–727
Ye D, Shin W-J, Li N, Tang W, Feng E, Li J, He P-L, Zuo J-P, Kim H, Nam K-Y, Zhu W, Seong B-L, No KT, Jiang H, Liu H (2012) Synthesis of C-4-modified zanamivir analogs as neuraminidase inhibitors and their anti-AIV activities. Eur J Med Chem 54:764–770
Rudrawar S, Dyason JC, Rameix-Welti MA, Rose FJ, Kerry PS, Russell RJ, van der Werf S, Thomson RJ, Naffakh N, von Itzstein M (2010) Novel sialic acid derivatives lock open the 150-loop of an influenza A virus group-1 sialidase. Nat Commun 1:113–117
von Itzstein M, Thomson R (2009) Anti-influenza drugs: the development of sialidase inhibitors. Handb Exp Pharmacol 189:111–154
Lew W, Chen X, Kim CU (2000) Discovery and development of GS 4104 (oseltamivir): an orally active influenza neuraminidase inhibitor. Curr Med Chem 7(6):663–672
Decarvalho C, Dafonseca M (2006) Carvone: why and how should one bother to produce this terpene. Food Chem 95(3):413–422
López G, Valencia A, Tress ML (2007) Firestar-prediction of functionally important residues using structural templates and alignment reliability. Nucleic Acids Res 35(Suppl2):573–577
Hariono M, Abdullah N, Damodaran KV, Kamarulzaman EE (2016) Potential new H1N1 neuraminidase inhibitors from ferulic acid and vanillin: molecular modelling, synthesis and in vitro assay. Nat Publ Gr https://doi.org/10.1038/srep38692
Wallace AC, Laskowski RA, Thornton JM (1995) LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions clean up structure. Protein Eng 8(2):127–134
Van Gunsteren FW, Billeter SR, Eising AA, Hünenberger PH, Krüger P, Mark AE, Scott WRP, Tironi IG (1996) Biomolecular simulation: the GROMOS96 manual and user guide. Vdf Hochschulverlag AG an der ETH Zürich, Zürich
Schüttelkopf AW, Van Aalten DMF (2004) PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr 60(8):1355–1363
Asadollahi-Baboli M, Mani-Varnosfaderani A (2013) Molecular docking, molecular dynamics simulation, and QSAR model on potent thiazolidine-4-carboxylic acid inhibitors of influenza neuraminidase. Med Chem Res 22(4):1700–1710
He JY, Li C, Wu G (2014) Discovery of potential drugs for human-infecting H7N9 virus containing R294K mutation. Drug Des Devel Ther 8:2377–2390
Kumari R, Kumar R, Lynn A (2014) G-mmpbsa -a GROMACS tool for high-throughput MM-PBSA calculations. J Chem Inf Model 54(7):1951–1962
Morris G, Goodsell D (1998) Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem 19:1639–1662
Morris GM, Huey R, Olson AJ (2008) UNIT using AutoDock for ligand-receptor docking. Curr Protocols Bioinform. https://doi.org/10.1002/0471250953.bi0814s24
Kang JW, Hwang HJ, Song KO, Choi WY, Byun KR, Kwon OK, Lee JH, Kim WW, Muchtaridi M, Aliyudin A, Holik HA (2003) Potential activity of some natural products compounds as neuraminidase inhibitors based on molecular docking simulation and in vitro test. J Korean Phys Soc 5(3):65–73
Wang T, Wade RC (2001) Comparative binding energy (COMBINE) analysis of influenza neuraminidase-inhibitor complexes. J Med Chem 44:961–971
Liu Z, Zhao J, Li W, Wang X, Xu J, Xie J, Tao K, Shen L, Zhang R (2015) Molecular docking of potential inhibitors for influenza H7N9. Comput Math Methods Med. 2015:1–9
Patil R, Das S, Stanley A, Yadav L, Sudhakar A (2010) Optimized hydrophobic interactions and hydrogen bonding at the target-ligand interface leads the pathways of drug-designing. PLoS One 5(8):e12029
Acknowledgments
SAH thanks Malaysia Ministry of Higher Education (MOHE) for Fundamental Research Grant Scheme (FRGS14-098-0339) and myBrain15 scholarship for NJ.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
ESM 1
(DOCX 429 kb)
Rights and permissions
About this article
Cite this article
Jusoh, N., Zainal, H., Abdul Hamid, A. et al. In silico study of carvone derivatives as potential neuraminidase inhibitors. J Mol Model 24, 93 (2018). https://doi.org/10.1007/s00894-018-3619-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-018-3619-6