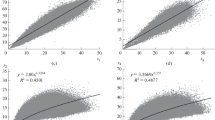
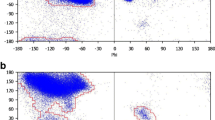
Abstract
Molecular and quantum mechanics calculations were carried out in a series of tripeptides (GXG, where X = D, N and C) as models of the unfolded states of proteins. The selected central amino acids, especially aspartic acid (D) and asparagine (N) are known to present significant average conformations in partially allowed areas of the Ramachandran plot, which have been suggested to be important in unfolded protein regions. In this report, we present the calculation of the propensity values through an umbrella sampling procedure in combination with the calculation of the NMR J-coupling constants obtained by a DFT model. The experimental NMR observations can be reasonably explained in terms of a conformational distribution where PPII and β basins sum up propensities above 0.9. The conformational analysis of the side chain dihedral angle (χ1), along with the computation of 3J(HαHβ), revealed a preference for the g − and g + rotamers. These may be connected with the presence of intermolecular H-bonding and carbonyl–carbonyl interactions sampled in the PPII and β basins. Taking into account all those results, it can be established that these residues show a similar behavior to other amino acids in short peptides regarding backbone φ,ψ dihedral angle distribution, in agreement with some experimental analysis of capped dipeptides.






Similar content being viewed by others
References
Shi ZS, Chen K, Liu ZG, Kallenbach NR (2006) Conformation of the backbone in unfolded proteins. Chem Rev 106(5):1877–1897. doi:10.1021/cr040433a
Beck DAC, Alonso DOV, Inoyama D, Daggett V (2008) The intrinsic conformational propensities of the 20 naturally occurring amino acids and reflection of these propensities in proteins. Proc Natl Acad Sci USA 105(34):12259–12264. doi:10.1073/pnas.0706527105
Grdadolnik J, Mohacek-Grosev V, Baldwin RL, Avbelj F (2011) Populations of the three major backbone conformations in 19 amino acid dipeptides. Proc Natl Acad Sci USA 108(5):1794–1798. doi:10.1073/pnas.1017317108
Jensen MR, Zweckstetter M, Huang J-R, Backledge M (2014) Exploring free-energy landscapes of intrinsically disordered proteins at atomic resolution using NMR spectroscopy. Chem Rev 114(13):6632–6660. doi:10.1021/cr400688u
Rybka K, Toal SE, Verbaro DJ, Mathieu D, Schwalbe H, Schweitzer-Stenner R (2013) Disorder and order in unfolded and disordered peptides and proteins: a view derived from tripeptide conformational analysis. II. Tripeptides with short side chains populating asx and beta-type like turn conformations. Prot Struct Funct Bioinf 81(6):968–983. doi:10.1002/prot.24226
Schweitzer-Stenner R, Hagarman A, Toal S, Mathieu D, Schwalbe H (2013) Disorder and order in unfolded and disordered peptides and proteins: a view derived from tripeptide conformational analysis. I. Tripeptides with long and predominantly hydrophobic side chains. Proteins 81(6):955–967. doi:10.1002/prot.24225
Ho BK, Thomas A, Brasseur R (2003) Revisiting the Ramachandran plot: hard-sphere repulsion, electrostatics, and H-bonding in the alpha-helix. Protein Sci 12(11):2508–2522. doi:10.1110/ps.03235203
Schweitzer-Stenner R (2012) Conformational propensities and residual structures in unfolded peptides and proteins. Mol BioSyst 8(1):122–133. doi:10.1039/c1mb05225j
Adzhubei AA, Sternberg MJE, Makarov AA (2013) Polyproline-II Helix in proteins: structure and function. J Mol Biol 425(12):2100–2132. doi:10.1016/j.jmb.2013.03.018
Toal S, Schweitzer-Stenner R (2014) Local order in the unfolded state: conformational biases and nearest neighbor interactions. Biomolecules 4(3):725–773. doi:10.3390/biom4030725
Bhowmick P, Guharoy M, Tompa P (2015) Bioinformatics approaches for predicting disordered protein motifs. In: Felli IC, Pierattelli R (eds) Intrinsically disordered proteins studied by NMR spectroscopy, vol 870. Advances in experimental medicine and biology. Springer, Berlin, pp 291–318. doi:10.1007/978-3-319-20164-1_9
Chou PY, Fasman GD (1978) Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol 47:45–148
Srinivasan N, Anuradha VS, Ramakrishnan C, Sowdhamini R, Balaram P (1994) Conformational characteristics of asparaginyl residues in proteins. Int J Pept Protein Res 44(2):112–122
Pal D, Chakrabarti P (2002) On residues in the disallowed region of the Ramachandran map. Biopolymers 63(3):195–206. doi:10.1002/bip.10051
Baldwin RL (2007) Energetics of protein folding. J Mol Biol 371(2):283–301. doi:10.1016/j.jmb.2007.05.078
Eker F, Griebenow K, Cao XL, Nafie LA, Schweitzer-Stenner R (2004) Preferred peptide backbone conformations in the unfolded state revealed by the structure analysis of alanine-based (AXA) tripeptides in aqueous solution. Proc Natl Acad Sci USA 101(27):10054–10059. doi:10.1073/pnas.0402623101
Oh K-I, Lee K-K, Park E-K, Jung Y, Hwang G-S, Cho M (2012) A comprehensive library of blocked dipeptides reveals intrinsic backbone conformational propensities of unfolded proteins. Prot Struct Funct Bioinf 80(4):977–990. doi:10.1002/prot.24000
Schweitzer-Stenner R (2006) Advances in vibrational spectroscopy as a sensitive probe of peptide and protein structure—a critical review. Vib Spectrosc 42(1):98–117. doi:10.1016/j.vibspec.2006.01.004
Karplus M (1959) Contact electron-spin coupling of nuclear magnetic moments. J Chem Phys 30(1):11–15. doi:10.1063/1.1729860
Deane CM, Allen FH, Taylor R, Blundell TL (1999) Carbonyl-carbonyl interactions stabilize the partially allowed Ramachandran conformations of asparagine and aspartic acid. Protein Eng 12(12):1025–1028. doi:10.1093/protein/12.12.1025
Bartlett GJ, Newberry RW, VanVeller B, Raines RT, Woolfson DN (2013) Interplay of hydrogen bonds and n ->pi* interactions in proteins. J Am Chem Soc 135(49):18682–18688. doi:10.1021/ja4106122
Cruz VL, Ramos J, Martinez-Salazar J (2012) Assessment of the intrinsic conformational preferences of dipeptide amino acids in aqueous solution by combined umbrella sampling/MBAR statistics. A comparison with experimental results. J Phys Chem B 116(1):469–475. doi:10.1021/jp206757j
Jorgensen WL, Tiradorives J (1988) The OPLS potential functions for proteins—energy minimizations for crystals of cyclic-peptides and crambin. J Am Chem Soc 110(6):1657–1666
Kaminski GA, Friesner RA, Tirado-Rives J, Jorgensen WL (2001) Evaluation and reparametrization of the OPLS-AA force field for proteins via comparison with accurate quantum chemical calculations on peptides. J Phys Chem B 105(28):6474–6487. doi:10.1021/jp003919d
Ravindranathan KP, Gallicchio E, Levy RM (2005) Conformational equilibria and free energy profiles for the allosteric transition of the ribose-binding protein. J Mol Biol 353(1):196–210. doi:10.1016/j.jmb.2005.08.009
Wang J, Gu Y, Liu H (2006) Determination of conformational free energies of peptides by multidimensional adaptive umbrella sampling. J Chem Phys 125(9):094907. doi:10.1063/1.2346681
Shirts MR, Chodera JD (2008) Statistically optimal analysis of samples from multiple equilibrium states. J Chem Phys 129(12):124105. doi:10.1063/1.2978177
Hess B, Kutzner C, van der Spoel D, Lindahl E (2008) GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput 4(3):435–447. doi:10.1021/ct700301q
Jorgensen WL, Tirado-Rives J (2005) Potential energy functions for atomic-level simulations of water and organic and biomolecular systems. Proc Natl Acad Sci USA 102(19):6665–6670. doi:10.1073/pnas.0408037102
Lawrence CP, Skinner JL (2003) Flexible TIP4P model for molecular dynamics simulation of liquid water. Chem Phys Lett 372(5–6):842–847. doi:10.1016/s0009-2614(03)00526-8
Kwac K, Lee KK, Han JB, Oh KI, Cho M (2008) Classical and quantum mechanical/molecular mechanical molecular dynamics simulations of alanine dipeptide in water: comparisons with IR and vibrational circular dichroism spectra. J Chem Phys 128(10):105106. doi:10.1063/1.2837461
Hess B, Bekker H, Berendsen HJC, Fraaije J (1997) LINCS: a linear constraint solver for molecular simulations. J Comput Chem 18(12):1463–1472
Berendsen HJC, Postma JPM, Vangunsteren WF, Dinola A, Haak JR (1984) Molecular-dynamics with coupling to an external bath. J Chem Phys 81(8):3684–3690
Darden T, York D, Pedersen L (1993) Particle mesh Ewald—an N.LOG(N) method for Ewald sums in large systems. J Chem Phys 98(12):10089–10092
Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG (1995) A smooth particle mesh Ewald method. J Chem Phys 103(19):8577–8593
Frisch GWT MJ, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA, Nakatsuji H, Caricato M, Li X, Hratchian HP, Izmaylov AF, Bloino J, Zheng G, Sonnenberg JL, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Vreven T, Montgomery JA Jr, Peralta JE, Ogliaro F, Bearpark M, Heyd JJ, Brothers E, Kudin KN, Staroverov VN, Kobayashi R, Normand J, Raghavachari K, Rendell A, Burant JC, Iyengar SS, Tomasi J, Cossi M, Rega N, Millam JM, Klene M, Knox JE, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Martin RL, Morokuma K, Zakrzewski VG, Voth GA, Salvador P, Dannenberg JJ, Dapprich S, Daniels AD, Farkas Ö, Foresman JB, Ortiz JV, Cioslowski J, Fox DJ (2009) Gaussian 09. Gaussian, Inc, Wallingford
Becke AD (1993) Density-functional thermochemistry. 3. The role of exact exchange. J Chem Phys 98(7):5648–5652. doi:10.1063/1.464913
Salvador P, Tsai IH, Dannenberg JJ (2011) J-coupling constants for a trialanine peptide as a function of dihedral angles calculated by density functional theory over the full Ramachandran space. Phys Chem Chem Phys 13(39):17484–17493. doi:10.1039/c1cp20520j
Tomasi J, Mennucci B, Cammi R (2005) Quantum mechanical continuum solvation models. Chem Rev 105(8):2999–3093. doi:10.1021/cr9904009
Cheeseman JR, Trucks GW, Keith TA, Frisch MJ (1996) A comparison of models for calculating nuclear magnetic resonance shielding tensors. J Chem Phys 104(14):5497–5509. doi:10.1063/1.471789
Deng W, Cheeseman JR, Frisch MJ (2006) Calculation of nuclear spin-spin coupling constants of molecules with first and second row atoms in study of basis set dependence. J Chem Theory Comput 2(4):1028–1037. doi:10.1021/ct600110u
Cruz V, Ramos J, Martinez-Salazar J (2011) Water-mediated conformations of the alanine dipeptide as revealed by distributed umbrella sampling simulations, quantum mechanics based calculations, and experimental data. J Phys Chem B 115(16):4880–4886. doi:10.1021/jp2022727
Degtyarenko IM, Jalkanen KJ, Gurtovenko AA, Nieminen RM (2007) l-Alanine in a droplet of water: a density-functional molecular dynamics study. J Phys Chem B 111(16):4227–4234. doi:10.1021/jp0676991
Jalkanen KJ, Degtyarenko IM, Nieminen RM, Cao X, Nafie LA, Zhu F, Barron LD (2008) Role of hydration in determining the structure and vibrational spectra of l-alanine and N-acetyl l-alanine N ′-methylamide in aqueous solution: a combined theoretical and experimental approach. Theor Chem Acc 119(1–3):191–210. doi:10.1007/s00214-007-0361-z
Kubelka J, Huang R, Keiderling TA (2005) Solvent effects on IR and VCD spectra of helical peptides: DFT-based static spectral simulations with explicit water. J Phys Chem B 109(16):8231–8243. doi:10.1021/jp0506078
Law PB, Daggett V (2010) The relationship between water bridges and the polyproline II conformation: a large-scale analysis of molecular dynamics simulations and crystal structures. Protein Eng Des Sel 23(1):27–33. doi:10.1093/protein/gzp069
Poon CD, Samulski ET, Weise CF, Weisshaar JC (2000) Do bridging water molecules dictate the structure of a model dipeptide in aqueous solution? J Am Chem Soc 122(23):5642–5643
Mobli M, Almond A (2007) N-Acetylated amino sugars: the dependence of NMR (3)J((H)(H)(N)(2))-couplings on conformation, dynamics and solvent. Org Biomol Chem 5(14):2243–2251. doi:10.1039/b705761j
Mackerell AD, Feig M, Brooks CL (2004) Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J Comput Chem 25(11):1400–1415. doi:10.1002/jcc.20065
Best RB, Buchete N-V, Hummer G (2008) Are current molecular dynamics force fields too helical? Biophys J 95(1):L07–L09. doi:10.1529/biophysj.108.132696
Hagarman A, Measey TJ, Mathieu D, Schwalbe H, Schweitzer-Stenner R (2010) Intrinsic propensities of amino acid residues in GxG peptides inferred from amide I′ band profiles and NMR scalar coupling constants. J Am Chem Soc 132(2):540–551
Harder E, Damm W, Maple J, Wu C, Reboul M, Xiang JY, Wang L, Lupyan D, Dahlgren MK, Knight JL, Kaus JW, Cerutti DS, Krilov G, Jorgensen WL, Abel R, Friesner RA (2016) OPLS3: a force field providing broad coverage of drug-like small molecules and proteins. J Chem Theory Comput 12(1):281–296. doi:10.1021/acs.jctc.5b00864
Lanza G, Chiacchio MA (2014) Ab initio MP2 and density functional theory computational study of AcAlaNH(2) peptide hydration: a bottom-up approach. ChemPhysChem 15(13):2785–2793. doi:10.1002/cphc.201402222
Chellgren BW, Miller A-F, Creamer TP (2006) Evidence for polyproline II helical structure in short polyglutamine tracts. J Mol Biol 361(2):362–371. doi:10.1016/j.jmb.2006.06.044
Jono R, Watanabe Y, Shimizu K, Terada T (2010) Multicanonical Ab inito QM/MM molecular dynamics simulation of a peptide in an aqueous environment. J Comput Chem 31(6):1168–1175. doi:10.1002/jcc.21401
Graf J, Nguyen PH, Stock G, Schwalbe H (2007) Structure and dynamics of the homologous series of alanine peptides: a joint molecular dynamics/NMR study. J Am Chem Soc 129(5):1179–1189. doi:10.1021/ja0660406
Kozminski W, Zhukov I, Pecul M, Sadlej J (2005) A protein backbone psi and phi angle dependence of (2)J(N(i)), C alpha(i-1): the new NMR experiment and quantum chemical calculations. J Biomol NMR 31(2):87–95. doi:10.1007/s10858-004-7563-7
Helgaker T, Jaszunski M, Ruud K, Gorska A (1998) Basis-set dependence of nuclear spin-spin coupling constants. Theor Chem Acc 99(3):175–182. doi:10.1007/s002140050321
Peralta JE, Scuseria GE, Cheeseman JR, Frisch MJ (2003) Basis set dependence of NMR spin-spin couplings in density functional theory calculations: first row and hydrogen atoms. Chem Phys Lett 375(5–6):452–458. doi:10.1016/s0009-2614(03)00886-8
Haehnke MJ, Richter C, Heinicke F, Schwalbe H (2010) The HN(COCA)HAHB NMR experiment for the stereospecific assignment of H-beta-protons in non-native states of proteins. J Am Chem Soc 132((3):918−+. doi:10.1021/ja909239w
Allen FH, Baalham CA, Lommerse JPM, Raithby PR (1998) Carbonyl–carbonyl interactions can be competitive with hydrogen bonds. Acta Crystallogr Sect B Struct Sci 54:320–329. doi:10.1107/s0108768198001463
Acknowledgments
Thanks are due to the CICYT (Ministerio de Economia y Competitividad) (Project MAT2012-36341) for financial support. This research was also funded by the CSIC (Consejo Superior de Investigaciones Científicas, Spain) under the Grant PIE-201360E097. J. R. acknowledges financial support through the Ramón y Cajal program (Contract RYC-2011-09585). The authors also thank the “Centro de Supercomputación de Galicia (CESGA)” and “Secretaria General Adjunta de Informatica (SGAI-CSIC)” for computational resources. We want to thank Prof. Schweitzer-Stenner, whose private comments some years ago motivated the present work. Computational support in the use of the distributed computing to the Ibercivis team is acknowledged (http://www.ibercivis.es). We are also very grateful to the anonymous citizens who have made available their desktop computers in an altruistic way.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Online Resource 1
Relative free energy map for GDG tripeptide using the Charmm27 force field. Energies are given in kT units and angles in degrees. (PDF 168 kb)
Online Resource 2
List of initial and final φ, ψ values corresponding to the fully optimized geometries for each tripeptide. The last column corresponds to the optimized electronic energies in atomic units. Figure showing the distribution of the optimized geometries in a Ramachandran plot (PDF 72 kb)
Online Resource 3
List of φ, ψ, χ 1 , and J-coupling constants calculated for the GDG case. Similar values were obtained for GNG. (PDF 97 kb)
Rights and permissions
About this article
Cite this article
Ramos, J., Cruz, V.L. Conformational analysis of short polar side-chain amino-acids through umbrella sampling and DFT calculations. J Mol Model 22, 273 (2016). https://doi.org/10.1007/s00894-016-3139-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-016-3139-1