Abstract
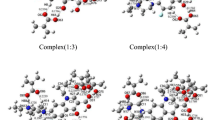
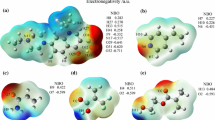
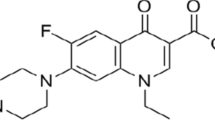
Recently, a series of computational and combinatorial approaches were employed to improve the efficiency of screening for optimal molecularly imprinted polymer (MIP) systems. In the present work, we investigated MIP systems based on enrofloxacin (ENRO) as the template molecule and either 2-vinyl-4,6-diamino-1,3,5-triazine (VDAT), 4-vinylpyridine (4-Vpy), acrylamide (AM), or trifluoromethacrylic acid (TFMAA) as the functional monomer. The optimized geometries of these systems, the optimal molar ratios of template to functional monomer, and the active sites in the systems were all identified using density functional theory (DFT) at the B3LYP/6-31G(d,p) level of theory. The imprinting mechanism was investigated by calculating the hydrogen nuclear magnetic resonance (1H NMR) spectra of the systems. The simulated results revealed that the MIP system corresponding to a 1:7 complex of TFMAA and ENRO contained the most H-bonds and presented the lowest (i.e., most negative) binding energy and the strongest interactions. MIPs of ENRO with the four functional monomers were prepared based on the optimal molar ratios of template to functional monomer determined in the simulations. Adsorption experiments suggested that TFMAA has the highest affinity (saturated adsorption 30.25 mg/g) among the four monomers for the template. Thus, we determined the optimal monomer and imprinting ratio for ENRO-imprinted MIPs and predicted their adsorption characteristics.

The preparation and extraction processes of MIPs with ENRO as template, TFMAA as functional monomer, and EDMA as cross-linker





Similar content being viewed by others
References
Ziółkowski H, Jaroszewski JJ, Maślanka T, Grabowski T, Katolik K, Pawęska J, Siemianowska M, Jasiecka A, Markiewicz W, Spodniewska A (2014) Res Vet Sci 97:99–104
Neves P, Berkane E, Gameiro P, Winterhalter M, de Castro B (2005) Biophys Chem 113:123–128
Babaahmady E, Khosravi A (2011) Afr J Pharm Pharmcol 5:2042–2045
Marchese S, Gentili A, Perret D (2005) TrAC Trend Anal Chem 24:704–733. doi:10.1016/j.trac.2005.02.007
Piacham T, Nantasenamat C, Isarankura-Na-Ayudhya C, Prachayasittikul V (2013) Excli J 12:701–718
Ebrahimzadeh H, Molaei K, Asgharinezhad AA, Shekari N, Dehghani Z (2013) Anal Chim Acta 767:155–162
Muhammad T, Cui L, Jide W, Piletska EV, Guerreiro AR, Piletsky SA (2012) Anal Chim Acta 709:98–104
Moreno-Bondi MC, Benito-Peña ME, Urraca JL, Orellana G (2012) Top Curr Chem 325:111–164. doi:10.1007/128_2010_94
Chianella I, Guerreiro A, Moczko E, Caygill JS, Piletska EV, De Vargas Sansalvador IMP, Whitcombe MJ, Piletsky SA (2013) Anal Chem 85:8462–8468
Granado VLV, Gutiérrez-Capitán M, Fernández-Sánchez C, Gomes MTSR, Rudnitskaya A, Jimenez-Jorquer (2014) Anal Chim Acta 809:141–147
Barkaline VV, Douhaya YV, Tsakalof A (2013) J Mol Model 19:359–369
Liu LK, Cao Y, Ma PF, Qiu CX, Xu WZ, Liu H, Huang WH (2014) RSC Adv 4:605–616
Cleland D, Olsson GD, Karlsson BCG, Nicholls IA, McCluskey A (2014) Org Biomol Chem 12:844–853
Liu JB, Shi Y, Tang SS, Jin RF (2015) J Sep Sci 38:1065–1071
Liu JB, Dai ZQ, Li B, Tang SS, Jin RF (2015) J Mol Model 20:2456–2465
Caro E, Marcé RM, Cormack PAG, Sherringtonb DC, Borrulla F (2006) Anal Chim Acta 562:145–151
Lu YK, Liu Y, Bian C, Lu GD, Qin XY (2009) CJI 11:26
Li XX, Bai LH, Wang H, Wang J, Huang YP, Liu ZS (2012) J Chromatogr A 1251:141–147
Wang YL, Liu JB, Tang SS, Chang HB, Liang DD (2013) Chem J Chin U 34:2880–2886. doi:10.7503/cjcu20130787
Lv YK, Zhang Q, Song YL, Yan SL (2011) Asian J Chem 23:4037–4041
Slinchenko O, Rachkov A, Miyachi H, Ogiso M, Minoura N (2004) Biosens Bioelectron 20:1091–1097
Ogiso M, Minoura N, Shinbo T, Shimizu T (2006) Biomaterials 27:4177–4182
Hoshina K, Horiyama S, Matsunaga H, Haginaka J (2011) J Pharma Biomed 55:916–922
Hiratsuka Y, Funaya N, Matsunaga H, Haginaka J (2013) J Pharma Biomed 75:180–185
Reddy SM, Hawkins DM, Phan QT, Stevenson D (2013) Sensor Actuators B 176:190–197
EL-Sharif HF, Hawkins DM, Stevenson D, Reddy SM (2014) Phys Chem Chem Phys 16:15483–15489
Subrahmanyam S, Guerreiro A, Poma A, Moczko E, Piletska E, Piletsky S (2013) Eur Polym J 49:100–105
Lata K, Sharma R, Naik L, Rajput YS, Mann B (2015) Food Chem 184:176–182
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Zakrzewski VG, Montgomery JA, Stratmann RE, Burant JC, Dapprich S, Millam JM, Daniels AD, Kudin KN, Strain MC, Farkas O, Tomasi J, Baroe V, Cossi M, Cammi R, Mennucci B, Pomelli C, Adamo C, Clifford S, Ochterski J, Petersson GA, Ayala PY, Cui Q, Morokuma K, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Cioslowski J, Ortiz JV, Baboul AG, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Gomperts R, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Gonzalez C, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Andres JL, Gonzalez C, Head-Gordon M, Replogle ES, Pople JA (2009) Gaussian 09, revision A.2. Gaussian Inc., Pittsburgh
Sastri VS, Perumareddi JR (1997) Corrosion 53:617–622
Parr RG, Szentpály LV, Liu SB (1999) J Am Chem Soc 121:1922–1924
Boys SF, Bernardi F (1970) Mol Phys 19:553–566
Houk KN (1975) Acc Chem Res 8:361–369
Dong WG, Yan M, Liu Z, Wu SG, Li YM (2007) Sep Purif Technol 53:183–188
Acknowledgments
The National Natural Science Foundation of China (no. 21302062), the Natural Science Foundation of Jilin Province (no. 201215180), and the Science and Technology Development Plan of Jilin Province (nos. 20130206099SF and 20150101018JC) are gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 5010 kb)
Rights and permissions
About this article
Cite this article
Dai, Z., Liu, J., Tang, S. et al. Optimization of enrofloxacin-imprinted polymers by computer-aided design. J Mol Model 21, 290 (2015). https://doi.org/10.1007/s00894-015-2836-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-015-2836-5