Abstract
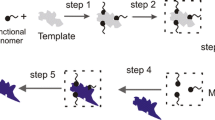
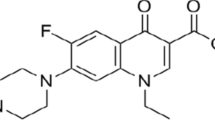
Molecularly imprinted polymers can be anticipated as synthetic imitation of natural antibodies, receptors and enzymes. In case of successful imprinting the selectivity and affinity of the imprint for substrate molecules are comparable with those of natural counterparts. The selection of the optimal functional monomer, monomer/template ratio as well as choosing of polymerization solvent is crucial determinants of the successful imprinting. In the present study the simulation approach to the development of molecular imprinting polymers for the extraction of new protein kinase ATP-competitive inhibitors is presented. By imprinting tri-O-acetyladenosine into polymer matrix the synthetic reproduction of adenosine triphosphate binding site to protein kinases can be fabricated and further used for adenosine triphosphate analogs screening in different sources. The optimized geometrical structure and energy of the pre-polymerization complexes of tri-O-acetyladenosine (template) with three different monomers—methacrylic acid, 3-vinyl benzoic acid and acrylamide in vacuum were calculated using hybrid quantum mechanical/molecular mechanical (QM/MM) approach. These calculations demonstrate that methacrylic acid forms the most stable complex with template, the next is 3-vinyl benzoic acid complex and the third—acrylamide one. The bond energies of the complexes are shown to increase monotonically as more monomers are linked to the template. The same conclusions are made from purely quantum self-consistent field calculations of pre-polymerization complex energy and structure. Hybrid calculation is shown to be effective and can substantially accelerate the development of the imprinting technology.

Pre-polymerization complex of MIP with tri-O-acetiladenosine template with 5 metacrylic acid monomers







Similar content being viewed by others
References
Lakka A, Mylonis I, Bonanou S, Simos G, Tsakalof A (2011) Isolation of hypoxia-inducible factor 1 (HIF-1) inhibitors from frankincense using a molecularly imprinted polymer. Invest New Drugs 29:1081–1089
Grant SK (2009) Therapeutic protein kinase inhibitors. Cell Mol Life Sci 66:1163–1177
Karin M (2005) Inflammation-activated protein kinases as targets for drug development. Proc Am Thorac Soc 2:386–390
Senn HM, Thiel W (2009) QM/MM methods for biomolecular systems. Angew Chem Int Ed 48:1198–1229
Farrington K, Regan F (2007) Investigation of the nature of MIP recognition: the development and characterisation of a MIP for Ibuprofen. Biosens Bioelectron 22:1138–1146
Jurema MW, Shields GC (1993) Ability of the PM3 quantum-mechanical method to model intermolecular hydrogen bonding between neutral molecules. J Comput Chem 14:89–104
Burshtein K, Shorygin P (1989) Quantum chemical calculations in organic chemistry and molecular spectroscopy. Nauka Publ, Moscow
Case DA, Cheatham TE 3rd, Darden T, Gohlke H, Luo R, Merz KM Jr et al. (2005) The Amber biomolecular simulation programs. J Comput Chem 26:1668–1688
Valiev M, Bylaska EJ, Govind N, Kowalski K, Straatsma TP, van Dam HJJ et al. (2010) NWChem: a comprehensive and scalable open-source solution for large scale molecular simulations. Comput Phys Commun 181:1477–1489
Smyth LA, Collins I (2009) Measuring and interpreting the selectivity of protein kinase inhibitors. J Chem Biol 2:131–151
http://www.nwchem-sw.org/index.php/QMMM_Parameters. Accessed 26 Jan 2011
Lattach Y, Archirel P, Remita S (2012) Influence of the chemical functionalities of a molecularly imprinted conducting polymer on its sensing properties: electromechanical measurement and semiempirical DFT calculations. J Phys Chem B 116:1467–1481
Hobza P, Mueller-Dethlefs K (2010) Non-covalent interactions: theory and experiment. Royal Society of Chemistry, Cambridge
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Barkaline, V.V., Douhaya, Y.V. & Tsakalof, A. Computer simulation based selection of optimal monomer for imprinting of tri-O-acetiladenosine in polymer matrix: vacuum calculations. J Mol Model 19, 359–369 (2013). https://doi.org/10.1007/s00894-012-1561-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-012-1561-6