Abstract
Treatment of C. difficile infection is one of the most difficult biomedical challenges. To develop novel antibacterials, researchers have been targeting bacterial molecular functions that are essential for its growth. The methionyl tRNA synthetase (MetRS) is strictly required for protein biosynthesis and success was reported in developing antibacterials to inhibit this enzyme. The present study was aimed at building and analyzing a homology model for C. difficile MetRS in the context of drug design. A homology model of C. difficile MetRS was constructed using Molecular Operating Environment (MOE) software. A. aeolicus MetRS was the main template while the query zinc binding domain was modeled using T. thermophilus MetRS. The model has been assessed and compared to its main template (Ramachandran, ERRAT and ProSA). The active site of the query protein has been predicted from its sequence using a detailed conservation analysis (ClustalW2). Using MOE software, suitable ligands were docked in the constructed model, including a C. difficile MetRS inhibitor REP3123 and the enzyme natural substrate, and the key active site residues and interactions were identified. These docking studies have validated the active site conformation in the constructed model and identified binding interactions.

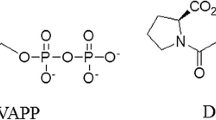
Final C. difficile Met-tRNA homology model and docking interaction of the inhibitor REP3123









Similar content being viewed by others
References
Richmond MA, Riggs MM, Eckstein BC, Donskey CJ (2008) Clostridium difficile infection in patients with SCI. J Spinal Cord Med 31:521–521
Gerding DN, Muto CA, Owens RC (2008) Treatment of Clostridium difficile infection. Clin Infect Dis 15:S32–42
Deaths involving Clostridium difficile: England and Wales 2004-2008 Office for National Statistics 19/8/2009. http://www.statistics.gov.uk
Deniziak MA, Barciszewski J (2001) Methionyl-tRNA synthetase. Acta Biochim Pol 48:337–350
Hountondji C, Dessen P, Blanquet S (1986) Sequence similarities among the family of aminoacyl-tRNA synthetases. Biochimie 68:1071–1078
Eriani G, Delarue M, Poch O, Gangloff J, Moras D (1990) Partition of transfer-RNA synthetases into 2 classes based on mutually exclusive sets of sequence motifs. Nature 347:203–206
Hurdle JG, O’Neill AJ, Chopra I (2005) Prospects for aminoacyl-tRNA synthetase inhibitors as new antimicrobial agents. Antimicrob Agents Chemother 49:4821–4833
Boyce JM (2001) MRSA patients: proven methods to treat colonization and infection. J Hosp Infect 48:S9–14
Jarvest RL, Berge JM, Berry V, Boyd HF, Brown MJ, Elder JS, Forrest AK, Fosberry AP, Gentry DR, Hibbs MJ, Jaworski DD, O’Hanlon PJ, Pope AJ, Rittenhouse S, Sheppard RJ, Slater-Radosti C, Worby A (2002) Nanomolar inhibitors of Staphylococcus aureus methionyl tRNA synthetase with potent antibacterial activity against gram-positive pathogens. J Med Chem 45:1959–1962
Finn J, Mattia K, Morytko M, Ram S, Yang Y, Wu X, Silverman J, Mak E, Gallant P, Keith D (2003) Discovery of a potent and selective series of pyrazole bacterial methionyl-tRNA synthetase inhibitors. Bioorg Med Chem Lett 13:2231–2234
Tandon M, Coffen D, Gallant P, Keith D, Ashwell M (2004) Potent and selective inhibitors of bacterial methionyl tRNA synthetase derived from an oxazolone-dipeptide scaffold. Bioorg Med Chem Lett 14:1909–1911
Finn J, Stidham M, Hilgers M, Kedar GC (2008) Identification of novel inhibitors of methionyl-tRNA synthetase (MetRS) by virtual screening. Bioorg Med Chem Lett 18:3932–3937
Critchley IA, Green LS, Young CL, Bullard JM, Evans RJ, Price M, Jarvis TC, Guilles JW, Janjic N, Ochsner U (2009) Spectrum of activity and mod of action of REP3123, a new antibiotic to treat Clostridium difficile. Antimicrob Agents Chemother 63:954–963
Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A (2003) ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res 31:3784-3788. www.expasy.org
Sebaihia M, Wren BW, Mullany P, Fairweather NF, Minton N, Stabler R, Thomson NR, Roberts AP, Cerdeño-Tárraga AM, Wang H, Holden MT, Wright A, Churcher C, Quail MA, Baker S, Bason N, Brooks K, Chillingworth T, Cronin A, Davis P, Dowd L, Fraser A, Feltwell T, Hance Z, Holroyd S, Jagels K, Moule S, Mungall K, Price C, Rabbinowitsch E, Sharp S, Simmonds M, Stevens K, Unwin L, Whithead S, Dupuy B, Dougan G, Barrell B, Parkhill J (2006) The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat Genet 38:779–786
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
RCSB Protein Data Bank (PDB) http://www.rcsb.org/pdb
Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, Gascuel O (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36:W465–469
Jones DT (1999) Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol 292:195–202
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) ClustalW and ClustalX version 2. Bioinformatics 23:2947–2948
Lobley A, Sadowski MI, Jones DT (2009) pGenTHREADER and pDomTHREADER: New Methods For Improved Protein Fold Recognition and Superfamily Discrimination. Bioinformatics 25:1761–1767
Molecular Operating Environment (MOE 2008.09) Chemical Computing Group Inc, Montreal Quebec Canada http://www.chemcomp.com. 2008.09
Weiner SJ, Kollman PA, Nguyen DT (1986) An all atom forcefield for simulations of proteins and nucleic acids. J Comput Chem 7:230–252
RAMPAGE Server http://ravenbioccam.ac.uk/rampage.php
UCLA-DOE Institute for Genomics & Proteomics Server http://www.doe-mbiucla.edu/Services
Wiederstein M, Sippl MJ (2007) ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res 35:W407–W410
Landes C, Perona JJ, Brunie S, Rould MA, Zelwar C, Steitz TA, Risler JL (1995) A structure-based multiple sequence alignment of all class I aminoacyl-tRNA synthetases. Biochimie 77:194–203
Sugiura I, Nureki O, Ugaji-Yoshikawa Y, Kuwabara S, Shimada A, Tateno M, Lorber B, Giege R, Moras D, Yokoyama S, Konno M (2000) Structure 8:197–208
Kamijo S, Fujii A, Onodera K, Wakabayashi K (2009) Analysis of conditions for KMSS loop in Tyrosyl-tRNA synthetase by building a mutant library. J Biochem 146:241–250
Spencer AC, Heck A, Takeuchi N, Watanabe K, Spremulli LL (2004) Characterization of the human mitochondrial methionyl-tRNA synthetase. Biochemistry 43:9743–9754
Crepin T, Schmitt E, Blanquet S, Mechulam Y (2004) Three-dimensional structure of methionyl-tRNA synthetase from Pyrococcus abyssi. Biochemistry 3:2635–2644
Mechulam Y, Schmitt E, Maveyraud L, Zelwer C, Nureki O, Yokoyama S, Konno M, Blanquet S (1999) Crystal structure of Escherichia coli methionyl-tRNA synthetase highlights species-specific features. J Mol Biol 294:1287–1297
Xu B, Krudy GA, Rosevear PR (1993) Identification of the metal ligands and characterization of a putative zinc finger in methionyl-tRNA synthetase. J Biol Chem 268:16259–16264
RNAfold server: http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi
Mathews DH, Sabina J, Zucker M, Turner H (1999) Expanded sequence dependence of thermodynamic parameters provides robust prediction of RNA secondary structure. J Mol Biol 288:911–940
Cavarelli J, Delagoute B, Eriani G, Gangloff J, Moras D (1998) L-arginine recognition by yeast arginyl-tRNA synthetase. EMBO J 17:5438–5448
Nureki O, Vassylyev DG, Katayanagi K, Shimizu T, Sekine S, Kigawa T, Miyazawa T, Yokoyama S, Morikawa K (1995) Architectures of class-defining and specific domains of glutamyl-tRNA synthetases. Science 267:1958–1965
Morales AJ, Swairjo MA, Schimmel P (1999) Structure-specific tRNA-binding protein from the extreme thermophile Aquifex aeolicus. EMBO J 18:3475–3483
Crepin T, Schmitt E, Mechulam Y, Sampson PB, Vaughan MD, Honek JF, Blanquet S (2003) Use of analogues of methionine and methionyl adenylate to sample conformational changes during catalysis in Escherichia coli methionyl-tRNA synthetase. J Mol Biol 332:59–72
Green LS, Bullard JM, Ribble W, Dean F, Ayers DF, Ochsner UA, Janjic N, Jarvis TC (2009) Inhibition of methionyl-tRNA synthetase by REP8839 and effects of resistance mutations on enzyme activity. Antimicrob Agents Chemother 53:86–94
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Al-Moubarak, E., Simons, C. A homology model for Clostridium difficile methionyl tRNA synthetase: active site analysis and docking interactions. J Mol Model 17, 1679–1693 (2011). https://doi.org/10.1007/s00894-010-0871-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-010-0871-9