Abstract
In this work we used a combination of classical molecular dynamics and simulated annealing techniques to shed more light on the conformational flexibility of 12 adenosine triphosphate (ATP) analogues in a water environment. We present simulations in AMBER force field for ATP and 12 published analogues [Shah et al. (1997) Proc Natl Acad Sci USA 94: 3565–3570]. The calculations were carried out using the generalized Born (GB) solvation model in the presence of the cation Mg2+. The ion was placed at a close distance (2 Å) from the charged oxygen atoms of the beta and gamma phosphate groups of the −3 negatively charged ATP analogue molecules. Analysis of the results revealed the distribution of inter-proton distances H8–H1′ and H8–H2′ versus the torsion angle ψ (C4–N9-C1′–O4′) for all conformations of ATP analogues. There are two gaps in the distribution of torsion angle ψ values: the first is between −30 and 30 degrees and is described by cis-conformation; and the second is between 90 and 175 degrees, which mostly covers a region of anti conformation. Our results compare favorably with results obtained in experimental assays [Jiang and Mao (2002) Polyhedron 21:435–438].

Dihedral O4′–C1′–N9–C4 angle dependence on inter-proton distances H8–H1′ (crosses) and H8–H2′ (dots) measured for ATP
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Adenosine triphosphate (ATP) is one of the most important molecules on Earth, present in all cells of all living organisms. This high-energy nucleotide powers, in several different ways, most biochemical processes that require energy. One such way is the transfer of a phosphate group to another molecule in a process called phosphorylation. This reaction is carried out by enzymes called kinases. Identification of substrates that are phosphorylated by specific kinases is difficult because of the enormous number of these enzymes, and also because kinases display overlapping substrate specificities [1, 2]. The approach presented by Shah et al. [3] is based on using both mutatated kinases that enlarge the ATP-binding pocket, and ATP analogues, whose specificity allows the kinase substrates to be identified. This method was used successfully to study Rous sarcoma virus tyrosine kinase [3].
The process of designing ATP-analogues complementary to modified kinases has to begin by understanding the conformational behavior of the nucleotide, and assurance that the modification introduced into ATP does not change its conformational properties. In this work, we study the conformation of the ATP molecule and the 12 analogues proposed by Shah et al. [3] bound with magnesium cation (Mg2+) using molecular dynamics (MD) simulation enhanced with simulated annealing (SA). We present a full set of AMBER force-field parameters for each of the ATP analogues, which provides the possibility to use models of these molecules in other computational experiments, such as docking and molecular modeling of the interaction between such analogues and kinases.
Since the discovery of protein kinase activity in 1954 [4], the field of protein kinase drug discovery has advanced dramatically. More and more researchers are involved in the design of new kinase inhibitors, as there is much focus on this subject by the pharmaceutical industry. Molecular modeling is one of the most helpful tools in this field. For example, molecular modeling was used successfully in studies on inhibitors of vascular endothelial growth factor receptor tyrosine kinase [5], the cyclin-dependent kinase family [6, 7], as well as in the case of the serine-threonine kinases p38 [8], Aurora A [9] or checkpoint kinase 1 [10]. The models presented in this work, together with their AMBER force-field parameters, can also be used for modeling kinase inhibitors as well as for designing ATP analogues other than those shown here.
Methods
Initial models
The ATP analogues considered in this work were taken from a set presented by Shah et al. [3]. Models of these ATP analogue molecules were built using MOLDEN [11], using also a model of the ATP molecule from the Structural Cambridge Database (entry ADENTP03 [12]) as a template. Two of the ATP analogue models, namely N6-methoxy ATP (AT1P) and N6-pyrrolidino ATP (AT7P) were built in our previous work [13]. Hybridizations of atom N6 in ATP-derivatives were determined by comparison with molecules having the N-substituent group attached to the aromatic ring. A comparison of crystal structures with the ATP-models is presented in Table 1.The charge of ATP and its analogues was −3, consistent with the models presented by Shah et al. [3]. Parameterization of the ATP analogues to the AMBER force field was performed as recommended in the AMBER [14] manual. Restrained electrostatic potential (RESP) was used to obtain partial atomic charges of ATP and its 12 analogues. The structures of the models were subjected to geometry optimization at the HF/6-31G* level using the ab-initio chemistry package GAMESS [15]. Charges were calculated from optimized geometries using R.E.D. (see http://q4md-forcefieldtools.org/RED/). A complete set of information on the assigned atom types and atomic charges in LEaP Object File Format (OFF) files can be found in the electronic supplementary material. All modifications introduced to the AMBER force field [16], which were used to parameterize ATP analogues, are shown in Table 2.
ATP requires the presence of a divalent cation, usually magnesium, which is coordinated by ATP phosphate groups. The presence of the ion is crucial to obtaining proper conformation of ATP but is also very important in catalyzing the phosphotransfer reaction [17], particularly in kinases [18]. One magnesium cation (Mg2+) was added to each model of ATP molecule using the LEaP command addIons. The addIons procedure places the counterion in a shell around one or more ATP analogue anion using a Coulombic potential on a grid.
The reason why only one magnesium cation was added can be found in our previous work [13, 19], where ATP and two ATP-derivatives respectively were docked to the serine-threonine kinase PrkC, and only one magnesium ion was found near the phosphates. An extensive study by Martinez et al. [20] on ATP conformations and ion binding modes in the active site of anthrax edema factor also supported a mechanism of kinase activity based on the presence of one magnesium cation in the active site of the enzyme.
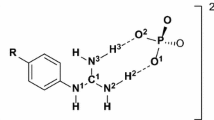
The magnesium cation was held using distance restraints during the SA simulations. To obtain an input distance restraint, we calculated the distances between the magnesium ion and four oxygen atoms of the phosphate groups beta and gamma (see Fig. 1), from initial structures after using addIons command. The values of distances varied between 2.0 and 4.5 Å. These values were used as boundary values and the force constants for distance restraints were 20 kcal mol−1 Å on each of the four distances (Mg–O1B, Mg–O2B, Mg–O1G and Mg–O2G).
Minimization and SA simulations
Initial structures of ATP and its analogues were optimized with SANDER, part of the AMBER 9.0 package [14] using Steepest Descent minimization for 5,000 steps, followed by the 15,000 steps of conjugate gradient minimization. Subsequently, all models were submitted to a 50-ps (50,000 steps with 1 fs time step) run of the SA protocol for 1,000 times. The single SA protocol consisted of four stages (see Fig. 2):
-
(1)
0–2 ps Short equilibration
-
(2)
2–10 ps heating from 300 K to 1,200 K
-
(3)
10–40 ps slow cooling from 1,200 K to 300 K
-
(4)
40–50 ps equilibration at 300 K
Each set of final coordinates from one SA simulation was used as a starting point for the following simulation. It is worth noting that there is a short equilibration period, in the first stage of the SA run, before the heating stage. Running the heating procedure immediately after minimization, brings instability to the system and causes unusually high energy.
The simulations were carried out using the implicit solvent model, namely the generalized Born (GB) solvation model developed by Onufriev, Bashford and Case (GBOBC) [21]. We are aware that the continuous solvent model is a crude approximation and does not take into account the molecular structure of water. It also neglects several modes of interaction, like hydrogen bond formation between solute atoms and water, but our results suggest that it is good enough to reproduce experimental results. The GB solvent model enabled us to perform quick SA simulations without causing artifacts in the water structure. For temperature regulation we used the Langevin thermostat [22] and a collision rate of 1 ps−1.
Analysis
Analysis of the ψ angle vs the inter-proton distance H8-H1′ and H8-H2′
The calculations of the torsion angle ψ, defined as O4′–C1′–N9′–C4′, and inter-proton distances (H8–H1′ and H8–H2′) were carried out using the PTRAJ module of AMBER 9 [14] based on the final structures from each of the simulation. Collected data are presented in Fig. 3 which was created using the gnuplot program (http://www.gnuplot.info). Points illustrating the dependence between the torsion angle (O4′–C1′–N9–C4) and inter-proton distances were fitted to the function: \( {f(x) = A{\hbox{*sin}}\left( {\left[ {x - o} \right]/B)} \right) + C} \) (Fig. 3a). Parameters obtained after the fitting procedure are presented in Table S1 (see electronic supplementary material). Fitting was done only for calculations concerning ATP, to visualize the shape of the plot of this function.
Dihedral O4′–C1′–N9–C4 angle dependence on inter-proton distances H8–H1′ (crosses) and H8–H2′ (dots) measured for ATP (a) and its analogues (b). This plot can be compared to the plot presented by Jiang et al. [24], the results of which were obtained from crystallography and NMR system (CNS) experiments
Results and discussion
Here we report the results of the conformational analysis carried out after MD simulations.
Magnesium cation restraints
The distances between magnesium and oxygen atoms (O) were measured to verify if the magnesium cation was kept in place by the restraints during our simulations. An illustration of what happens during a single SA run is presented in Fig. 1. We show only the first run for ATP, because the remaining runs for the ATP derivatives are very similar and do not show any significant variations.
Figure 1 shows that the distances between the Mg2+ cation and the four charged oxygen atoms vary between 2.0 Å and 4.5 Å. The restraints hold the magnesium cation at the imposed distances. The Mg2+ cation is free to move within the restraints, e.g., around 25 ps, Mg2+ moved from oxygen O2B towards O1B and then went back to its previous position.
To better understand what is happening in the same time scale, Fig. 2 illustrates the time/temperature dependency during the same, single SA run. Significant changes in the four monitored distances were observed at the slow cooling stage of the SA run, namely between 10 ps and 40 ps. This is understandable, because the slow cooling stage allows the system to obtain the most ordered or lowest energy arrangement of molecules, and stay close to the equilibrium state [23].
To verify if the magnesium cation is kept close to the initial position during all 1,000 SA MD runs, we measured the distances between Mg2+ and the charged oxygen atoms from the beta and gamma phosphate groups (O1B, O2B, O1G and O2G) for each snapshot taken at the end of every SA MD run.
Figure 4 presents the frequencies of occurrence of the distance values around 2.0 Å and 4.5 Å for ATP (ATP-analogues showed similar results). The frequency of occurrence of values at 2.0 Å is higher than the frequency of occurrence of values at 4.5 Å. We observed that when the cation is bound within a close distance (2.0 Å) between two oxygen atoms it does not change its position, which is why we observe the Mg2+ cation at almost the same distances. Close distance also represents stronger ion interaction between the magnesium cation and the negatively charged oxygen atoms. Occurrences of distance values in the range between 2 Å and 3.5 Å are very rare because this position is energetically unfavorable.
Analysis of the distribution of the values of torsion angle O4′–C1′–N9–C4
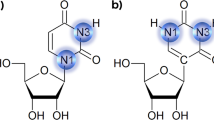
ATP conformation can be partially defined by value of the torsion angle between atoms O4′–C1′–N9–C4 (see Fig. S1). If the value varies between 0 ± 90 degrees, then the conformation is denoted as “syn”, and if the value varies between 180 ± 90 degrees than the conformation is denoted as “anti”. We checked the distribution of values in conformations of our models, which were submitted to SA MD. Values of the dihedral angle (O4′–C1′–N9–C4) were measured for each of the snapshots taken after the equilibration stage. Table 3 presents the measured values of the torsion angle for each of the ATP-analogues. As an example of the distribution of the analyzed dihedral angle, Fig. 5 shows the distribution of values of the O4′–C1′–N9–C4 angle in ATP.
Both Table 3 and Fig. 5 clearly show that there are two maxima in the frequency of occurrence of the torsion angle value. The first maximum is present around −120 degrees (conformation anti) and the second maximum is around 60 degrees (conformation syn). The ATP molecule or its analogue frequently adopts the anti conformation when it binds inside the ATP-binding pocket. The results show that the first maximum is broad and the other is rather sharp. These two maxima represent two major sets of conformations of ATP and its analogues, and the transitions between them may be essential for kinase activation.
Dependence between O4′–C1′–N9–C4 angle and inter-proton distances H8–H1′/H8–H2′
The distribution of conformations of ATP can also be partially described using analysis of the inter-proton distances H8–H1′ and H8–H2′ versus the torsion angle ψ (C4–N9–C1′–O4′). This analysis was previously applied by Jiang and Mao [24] to the interpretation of combined NMR and molecular modeling experiments on unmodified ATP molecules. Comparison of our results and those obtained by Jiang and Mao shows that we observe the same structural behavior. From Fig. 3, we can clearly conclude that there are two main conformations of ATP model, the first, which is mostly in the anti conformation with ψ angle value of −120 ± 60 degrees ,and the second (syn) with ψ angle value of 45 ± 30 degrees. The two least frequent conformations are characterized by ψ angle values of −30 ± 30 and 120 ± 30, respectively.
Dihedral angle C4–N9–C1′–O4′ analysis shows that, in ATP as well as in the 12 analogues, there are two gaps in the distribution of angle values ranges, the first between −30 and 30 degrees described by cis-conformation, and the second between 90 and 175 degrees, which mostly covers region of anti conformation, as presented on the right side of Fig. 3a. The cis-conformation is forbidden only for some of the ATP analogue models, namely 3, 7, 10 and 12 (see Fig. 3b). This is a transitional conformation between the “inactive conformation” (ψ angle values around 60) and the “active conformation” (ψ angle values range around 160). By the “active conformation” we mean a conformation of ATP that is able to form a complex with protein. If the molecular conformation falls into the inactive state and the cis-conformation is forbidden, then there is no possibility to change the conformation into the active state. This conformational “gap” can possibly decrease the binding affinity of such a ligand. This hypothesis explains the results of biological experiments, namely the low values of inhibition for ATP-analogues 3, 7 and 10 found by Shokat and coworkers [3, 25].
The second region is forbidden for all the ligands, ATP and its derivatives. To understand why the second region is forbidden for all the ATP models, we calculated the energy profile of the ATP model, changing the torsion angle by rotating the adenine ring. We observed an increase in Van der Waals (VDW) energy in the second forbidden-conformational state (data not shown). Close distances between the adenine nitrogen atom N9 and hydrogen H2′ and H3′ atoms from ribose ring were revealed by visual inspection of the conformation.
To summarize, the anti-conformation is disallowed due to steric effects in all of the models, while in contrast the presence of the cis-conformation is crucial for biological activity.
How does ATP conformation depend on the conformation of sugar?
The conformational change between the active and inactive stage is possible only through the cis-conformation. The role of ribose conformation might also be significant in obtaining the active/inactive stage. To verify this hypothesis, we calculated the relationship of ψ angle and torsion angle between atoms C1′–C2′–C3′–C4′, which defines the sugar conformation (see Fig. S1 and Fig. 6). The most commonly populated sugar conformations [26] were the C3′-endo region, corresponding to a torsion angle around 35°, and the C2′-endo region with torsion angle values around −35°.
Dihedral O4′–C1′–N9–C4 angle dependence on sugar conformation, described by dihedral C1′–C2′–C3′–C4′angle measured for ATP (a) and its analogues (b). In a, the schematic conformational transition from inactive to active state is also shown. The conformations were not clustered but we indicate only those that were close to each other (black boxes in a)
We observed that rotation of the adenine depends on the conformation of the sugar. Figure 6 presents four main conformations, two in the active state and two in the inactive state. One of the conformations in the inactive state is preferable and reflects the C2′-endo sugar conformation and the torsion ψ angle around 45 degrees. For most of the analogues it is not possible to change the adenine position from the inactive to the active state having the C2′-endo conformation. Furthermore, the switch between the active and inactive state is possible with the C3′-endo sugar conformation present. The same conformational behavior was observed in the case of the ATP molecule. Focusing on the active conformation, we observed that it is also favorable when the sugar is in the C2′-endo conformation.
We speculate that changes in sugar conformation are involved in the transition from the inactive to the active state of ATP analogues, and that the sequence of changes includes the following stages (Fig. 6a):
-
(5)
C2′-endo cis-conformation inactive
-
(1)
C3′-endo cis-conformation
-
(2)
C3′-endo anti-conformation
-
(3)
C2′-endo anti-conformation active
Summary
In this work we present parameters introduced into the AMBER force field and MD calculations for ATP and 12 ATP analogues proposed by Shah et al. [3] with one magnesium cation, Mg2+. The ion was found close (2 Å) to the charged oxygen atoms of the beta and gamma phosphate groups of the ATP analogue molecule. Analyzing the O4′–C1′–N9–C4 dihedral angle values, which partially describe the conformation of ATP (and its N6-substituted derivatives), we discovered the existence of two maxima. One sharp maximum located near the +60° value of the angle, which we call the “inactive-state”; and a second, broad maximum located close to −120°, called the “active-state”. The active-state conformation is frequently found when ATP is bound in the ATP-binding pocket of kinases. Change between the states is possible only through cis-conformation (0 ± 30°), which is not allowed for all ATP-analogues, namely AT3P, AT7P, AT10P and AT12P. These results correspond to the findings of Shah et al. [3], which explained the low values of inhibition activity of those ATP-derivatives with wild-type and mutant Rous sarcoma tyrosine kinase. Additionally, we determined the character of the contribution of the conformation of the ribose ring to the transition between the inactive/active state. The path from the inactive state to the active state leads through changes between the C2′-endo and C3′-endo of the sugar.
Substitutions proposed by Shah et al. on the N6 adenine atom of ATP do not influence the main conformational properties of the nucleoside. Parameters introduced by us to the AMBER force field tested by MD simulations gave reasonable results regarding conformation, which are comparable to experimental values [24].
Abbreviations
- ATP:
-
Adenosine triphosphate
- MD:
-
Molecular dynamics
- SA:
-
Simulated annealing
- A*TP:
-
Adenosine triphosphate derivatives
- RESP:
-
Restrained electrostatic potential
- GB:
-
Generalized Born
- VDW:
-
Van der Waals
References
Hunter T (1995) Protein kinases and phosphatases: the yin and yang of protein phosphorylation and signaling. Cell 80:225–236
Songyang Z, Carraway KL 3rd, Eck MJ, Harrison SC, Feldman RA, Mohammadi M, Schlessinger J, Hubbard SR, Smith DP, Eng C et al (1995) Catalytic specificity of protein-tyrosine kinases is critical for selective signalling. Nature 373:536–539
Shah K, Liu Y, Deirmengian C, Shokat KM (1997) Engineering unnatural nucleotide specificity for Rous sarcoma virus tyrosine kinase to uniquely label its direct substrates. Proc Natl Acad Sci USA 94:3565–3570
Burnett G, Kennedy EP (1954) The enzymatic phosphorylation of proteins. J Biol Chem 211:969–980
Du J, Lei B, Qin J, Liu H, Yao X (2009) Molecular modeling studies of vascular endothelial growth factor receptor tyrosine kinase inhibitors using QSAR and docking. J Mol Graph Model 27:642–654
Rossi KA, Markwalder JA, Seitz SP, Chang CH, Cox S, Boisclair MD, Brizuela L, Brenner SL, Stouten PF (2005) Understanding and modulating cyclin-dependent kinase inhibitor specificity: molecular modeling and biochemical evaluation of pyrazolopyrimidinones as CDK2/cyclin A and CDK4/cyclin D1 inhibitors. J Comput Aided Mol Des 19:111–122
Perez PC, Caceres RA, Canduri F, Filgueira de Azevedo JW (2009) Molecular modeling and dynamics simulation of human cyclin-dependent kinase 3 complexed with inhibitors. Comput Biol Med 39:130–140
Ravindra GK, Achaiah G, Sastry GN (2008) Molecular modeling studies of phenoxypyrimidinyl imidazoles as p38 kinase inhibitors using QSAR and docking. Eur J Med Chem 43:830–838
Talele TT, McLaughlin ML (2008) Molecular docking/dynamics studies of Aurora A kinase inhibitors. J Mol Graph Model 26:1213–1222
Chen XM, Lu T, Lu S, Li HF, Yuan HL, Ran T, Liu HC, Chen YD (2009) Structure-based and shape-complemented pharmacophore modeling for the discovery of novel checkpoint kinase 1 inhibitors. J Mol Model 16:1195–1204
Schaftenaar G, Noordik JH (2000) Molden: a pre- and post-processing program for molecular and electronic structures. J Comput Aided Mol Des 14:1231–1234
Sugawara Y, Iwasaki H (1984) Crystal transformation and conformational change of disodium adenosine 5′-triphosphate and the structure of ATP Na2.2H2O. Acta Crystallogr Sec A 40:C68
Gruszczyński P, Kaźmierkiewicz R, Obuchowski M, Lammek B (2007) Theoretical modeling of PrkCc, serine-threonine protein kinase intracellular domain, complexed with ATP derivatives. QSAR Comb Sci 27:437–444
Case DA, Cheatham TE 3rd, Darden T, Gohlke H, Luo R, Merz KM Jr, Onufriev A, Simmerling C, Wang B, Woods RJ (2005) The Amber biomolecular simulation programs. J Comput Chem 26:1668–1688
Schmidt M, Baldridge K, Boatz J, Elbert S, Gordon M, Jensen J, Koseki S, Matsunaga N, Nguyen K, Su S, Windus T, Dupuis M, Montgomery J (1993) General atomic and molecular electronic structure system. J Comput Chem 14:1347–1363
Meagher KL, Redman LT, Carlson HA (2003) Development of polyphosphate parameters for use with the AMBER force field. J Comput Chem 24:1016–1025
Williams NH (2000) Magnesium ion catalyzed ATP hydrolysis. J Am Chem Soc 122:12023–12024
Lin X, Ayrapetov MK, Sun G (2005) Characterization of the interactions between the active site of a protein tyrosine kinase and a divalent metal activator. BMC Biochem 6:25
Gruszczyński P, Obuchowski M, Kaźmierkiewicz R (2010) Phosphorylation and ATP-binding induced conformational changes in the PrkC Ser/Thr kinase from B. subtilis. J Comput Aided Mol Des. doi:10.1007/s10822-010-9370-4
Martinez L, Laine E, Malliavin TE, Nilges M, Blondel A (2009) ATP conformations and ion binding modes in the active site of anthrax edema factor: a computational analysis. Proteins 77:971–983
Bashford D, Case DA (2000) Generalized born models of macromolecular solvation effects. Annu Rev Phys Chem 51:129–152
Pastor RW, Brooks BR, Szabo A (1988) An analysis of the accuracy of Langevin and molecular dynamics algorithms. Mol Phys 65:1409–1419
Durand MD, White SR (2000) Trading accuracy for speed in parallel simulated annealing with simultaneous moves. Parallel Comput 26:135–150
Jiang L, Mao X-A (2002) Conformation of adenosine-5′-triphosphate in the presence of Mg2+ at different pH. Polyhedron 21:435–438
Liu Y, Shah K, Yang F, Witucki L, Shokat KM (1998) Engineering Src family protein kinases with unnatural nucleotide specificity. Chem Biol 5:91–101
Altona C, Sundaralingam M (1972) Conformational analysis of the sugar ring in nucleosides and nucleotides. J Am Chem Soc 94:8205–8212
Allen FH (2002) The Cambridge Structural Database: a quarter of a million crystal structures and rising. Acta Crystallogr B 58:380–388
Lin LZ, Hu SF, Cordell GA (1996) 19[alpha]-hydroxygelsamydine from Gelsemium elegans. Phytochemistry 43:723–726
Wasserman HJ, Ryan RR, Layne SP (1985) Structure of acetanilide (C8H9NO) at 113 K. Acta Crystallogr C 41:783–785
Yang L, Fang H, Zheng H, Weng LL (2007) (3S)-4-Phenyl-3-(phenylaminooxy)butan-2-one. Corrigendum. Acta Cryst E63:e27, doi:10.1107/S1600536807041785
Silversides JD, Sparke AE, Archibald SJ (2006) N-(4-Nitrobenzyl)benzene-1, 2-diamine. Acta Cryst E62:o5944–o5946
Hokelek T, Kilic E, Tuzun C (1991) Structural investigations of benzo[c]cinnoline derivatives. II. Structures of 2-pyrrolidinobenzo[c]cinnoline and 4 pyrrolidinobenzo[c]cinnoline. Acta Crystallogr C 47:373–376
Liu X, Kassiou M, Christie MJ, Hambley TW (2001) Trishomocubanes: requirements for α receptor binding and subtype selectivity. Aust J Chem 54:31–36
James BD, Mutrofin S, Skelton BW, White AH (2003) 4-(piperidin-1-yl)pyridinium hexafluorophosphate at 150 K. Acta Crystallogr C 59:o622–o624
Tanaka N, Ashida T, Sasada Y, Kakudo M (1967) The crystal structure of riboflavin hydrobromide monohydrate. Bull Chem Soc Jpn 40:1739
Momiyama N, Yamamoto H (2002) Lewis acid promoted, O-selective, nucleophilic addition of silyl enol ethers to N=O bonds. Angew Chem Int Ed Engl 41:2986–2988
Acknowledgment
We would like to acknowledge Dr. Artur Sikorski for accessing the Cambridge Structural Database and for his valuable help during the research.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Gruszczyński, P., Smalara, K., Obuchowski, M. et al. ATP and its N6-substituted analogues: parameterization, molecular dynamics simulation and conformational analysis. J Mol Model 17, 1081–1090 (2011). https://doi.org/10.1007/s00894-010-0808-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-010-0808-3