Abstract
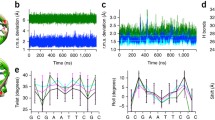
A software algorithm has been developed to investigate the folding process in B-DNA structures in vacuum under a simple and accurate force field. This algorithm models linear double stranded B-DNA sequences based on a local, sequential minimization procedure. The original B-DNA structures were modeled using initial nucleotide structures taken from the Brookhaven database. The models contain information at the atomic level allowing one to investigate as accurately as possible the structure and characteristics of the resulting DNA structures. A variety of DNA sequences and sizes were investigated containing coding and non-coding, random and real, homogeneous or heterogeneous sequences in the range of 2 to 40 base pairs. The force field contains terms such as angle bend, Lennard-Jones, electrostatic interactions and hydrogen bonding which are set up using the Dreiding II force field and defined to account for the helical parameters such as twist, tilt and rise. A close comparison was made between this local minimization algorithm and a global one (previously published) in order to find out advantages and disadvantages of the different methods. From the comparison, this algorithm gives better and faster results than the previous method, allowing one to minimize larger DNA segments. DNA segments with a length of 40 bases need approximately 4 h, while 2.5 weeks are needed with the previous method. After each minimization the angles between phosphate–oxygen-carbon A 1, the oxygen–phosphate–oxygen A 2 and the average helical twists were calculated. From the generated fragments it was found that the bond angles are A 1=150°±2°and A 2=130°±10°, while the helical twist is 36.6°±2° in the A strand and A 1=150°±6° and A 2=130±6° with helical twist 39.6°±2° in the B strand for the DNA segment with the same sequence as the Dickerson dodecamer.
Figure The final minimized DNA segment of the Dickerson dodecamer sequence represented by ball drawings and viewed (left) perpendicular and (right) down the helical axis










Similar content being viewed by others
References
Tidor B, Irikura KK, Brooks BR, Karplus M (1983) J Biomol Struct Dyn 1:231–252
Levitt M (1983) Cold Spring Harbor Symp Quant Biol 45:251–262
Rappe AK, Cassewit CJ (1997) Molecular mechanics across chemistry. University Science Books
Swamminathan S, Ravishaanker G, Beveridge DL (1991) J Am Chem Soc 113:5027-5040
Drukker K, Schatz GC (2000) J Phys Chem B 26:6108–6111
Bruant N, Flatters D, Lavery R, Genest D (1999) Biophys J 5:2366–2376
Jian H, Vologodski AV (1997) J Comput Phys 136:168–179
(a) Lavery R, Lebroun A (1999) Genetica 106:1–2; (b) Lavery R, Lebroun A (1999) Genetica 106:75–84
Olson WK, Zhurkin VB (2000) Curr Opin Struct Biol 3:286–297
Vlahovicek K, Pongor S (2000) Bioinformatics 11:1044–1045
Treger M, Westhof E (1987) J Mol Graphics 5:178–183
Srinivasan AR, Olson WK (1987) J Biomol Struct Dyn 4:895–938
Schlick T (1995) Curr Opin Struct Biol 5:245–262
MacKerrell Jr AD, Banavali N, Foloppe N (2000) Biopolymers 56:257–265
Schneider B, Neidle S, Berman HM (1997) Biopolymers 42:113-124
Sherer EC, Harris SA, Soliva R, Orozco M, Laughton CA (1999) J Am Chem Soc 121:5981–5991
Gabb HA, Prevost C, Bertucat G, Robert CH, Lavery R (1997) J Comput Chem 16:2001-2011
Duan Y, Wilkosz P, Crowley M, Rosenberg JM (1997) J Mol Biol 4:553–572
Ayadi L, Coulombeau C, Lavery R (1999) Biophys J 6:3218–3226
Lafontaine L, Lavery R (2000) Biophys J 2:680–685
Westcott TP, Tobias I, Olson WK (1997) J Chem Phys 10:3967–3980
Ramachandran G, Scchlick T (1995) Phys Rev E 6:6188–6203
Schlick T, Olson WK (1992) J Mol Biol 223:1089–1119
Schlick T, Olson WK (1992) Science 257:1110–1115
Ezaz-Nikpay K, Verdine GL (1992) J Am Chem Soc 114:6562–6563
Tereshko V, Minasov G, Egli M (1999) J Am Chem Soc 121:470–471
Tereshko V, Minasov G, Egli M (1999) J Am Chem Soc 121:6970–6970
Sfyrakis K, Provata A, Povey DC, Howlin BJ (2003) Molecular Simulations 29:555–575
Creighton TE (1993) Structures and Molecular Properties, 2nd edn. Proteins, New York
Goodfellow JM, Moss DS (1992) Computer modeling of bimolecular processes. Ellis Horwood
Voet D, Voet JG (1995) Biochemistry, 2nd edn. Wiley, New York
Olson WK, Srinivasan AR, Hao HH, Nauss JL (1988) In: Olson WK, Sarma MH, Sarma RH, Sundaralingman M (eds) Structure and expression 3, DNA bending and curvature. Adenine Press, New York, pp 225–242
Schurr JM (1985) Biopolymers 24:1233–1246
Selepova P, Kypr J (1985) Biopolymers 24:867–882
Shimada J, Yamakawa H (1985) J Mol Biol 184:319–329
Tsuru H, Wadati M (1986) Biopolymers 25:2083–2096
Tan RK, Harvey SC (1989) J Mol Biol 205:573-591
Provata A, Tsakiroglou M, Almirantis Y (2000) Non Randomness and Non Linearities in DNA. Proceedings to 1st Interdisciplinary Symposium on Non-linear Problems, Athens
Almirantis Y, Provata A (1999) J Stat Phys 97:233–262
Press WH, Teukolsky SA, Vetterling WT, Flannery BP (1992) Numerical Recipes in Fortran. The art of scientific computing, 2nd edn. Cambridge University Press, Cambridge
Mayo SL, Olafson BD, Goddard WA (1990) J Phys Chem 94:8897–8909
Sayle R (1994) Rasmol home pagehttp://www.rasmol.com
Albert B, Bray D, Lewis J, Raff M, Roberts K, Watson JD (1994) Molecular biology of the cell, 3rd edn. Garland Publishing
Zimmerman SB (1982) Annu Rev Biochem 51:395–427
Nunn CM, Jenkins TC, Neidle SE (1994) Eur J Biochem 226:953–961
Denisov AY, Zamaratski EV, Maltseva TV, Sandstrom A, Bekiroglou S, Altmann K–H, Egli M, Chattopadhyaya J (1998) J Biomol Struct Dyn 3:547–568
Maltseva TV, Altmann K–H, Egli M, Chattopadhyaya J (1998) J Biomol Struct Dyn 3:569–578
Ikeda H, Fermandez R Wilk A, Barchi Jr JJ, Huang X, Marquez VE (1998) Nucleic Acid Res 9:2237–2244
Gelasco A, Lippard SJ (1998) Biochemistry 26:9230–9239
Protein Data Bank Contents Guide. Atomic co-ordinates entry format description,http://www.rcsb.org
IUPAC: International Union of Pure and Applied Chemistry,http://:www.chem.qmul.ac.uk/iupac
Weiner SJ, Kollman PA, Case DA, Singh UC, Ghio C, Alagona G, Profeta S, Weiner P (1984) J Am Chem Soc 106:765–784
Author information
Authors and Affiliations
Corresponding author
Appendices
Appendix A
Initial structures are shown in Tables 6 and 7.
Appendix B
The average starting and final energy values of the minimized double B-DNA strands are shown in Table 8.
Rights and permissions
About this article
Cite this article
Sfyrakis, K., Provata, A., Povey, D.C. et al. Local sequential minimization of double stranded B-DNA using Monte Carlo annealing. J Mol Model 10, 185–197 (2004). https://doi.org/10.1007/s00894-004-0182-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-004-0182-0