Abstract
Biological soil crusts of extreme habitats (semi-deserts and deserts) are dominated by cyanobacteria and microalgae. The most abundant taxa are green algae belonging to the classes Chlorophyceae and Trebouxiophyceae. Specimens with sarcinoid-like morphology (cells arranged in packages) represent one group of these microalgae. The genus Pleurastrosarcina consists of two species, which were originally described as Chlorosarcina (P. brevispinosa and P. longispinosa). Both species are exclusively found from arid soils. However, these species were only reported few times and probably overlooked especially if no akinetes were present. During studying soil samples collected from different regions of the Atacama desert (Chile), we isolated two strains, which were morphologically similar to both Pleurastrosarcina species. The phylogenetic analyses confirmed that they belong to this genus. The ITS-2/CBC approach revealed that both new isolates represent a new species, P. terriformae. The comparison with other available strains demonstrated that this new species is not restricted to South America and was also found in coastal area in Europe. The six investigated strains showed a high phenotypic plasticity, which is reflected in the descriptions of several varieties.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Microalgae and cyanobacteria, which often form biological soil crusts, play an important role in arid regions such as semi-deserts and deserts around the world (Büdel et al. 2016 and references therein). Many new genera and species of coccoid green algae isolated from desert soil samples were described (i.e., Fučiková et al. 2014; Darienko and Pröschold 2019). Lewis and Flechtner (2002), Lewis and Lewis (2005) and Flechtner et al. (2013) studied the biodiversity of eukaryotic microalgae from different desert regions in North America. Büdel et al. (2009) investigated the biodiversity of southern African soil crusts. In all these studies, the dominant microalgal group were taxa belonging to two classes of green algae, Chlorophyceae and Trebouxiophyceae. As these investigations have shown, green algae with sarcinoid-like morphology often occur in biological soil crusts. They were classified as Chlorosarcinopsis (cell packages surrounded by mucilage and parietal chloroplasts with pyrenoids), Chlorosarcina (cell packages without mucilage and parietal chloroplasts without pyrenoids), or Desmochloris (cell packages without mucilage and parietal chloroplasts with pyrenoids). Detailed morphological descriptions about Chlorosarcinopsis, Chlorosarcina, and Desmochloris are summarized in Groover and Bold (1969), Chantanachat and Bold (1962), and Darienko et al. (2009), respectively. Phylogenetic analyses of several Chlorosarcinopsis and Desmochloris species have shown that they belong to the Chlorophyceae (Watanabe et al. 2006) and the Ulvophyceae (Darienko and Pröschold 2009, 2017), respectively. About the genus Chlorosarcina, very little is known, because only three species are available in public culture collections and only one species has been investigated in detail. Chlorosarcina stigmatica originally described by Deason (1959) was later transferred to Desmotetra based on ultrastructural investigations of the flagellar apparatus (Deason and Floyd 1987). The other two species of Chlorosarcina, C. brevispinosa and C. longispinosa, were not studied using an integrative approach.
Both species were described by Chantanachat and Bold (1962) from soil of semi-desert in Arizona and Moab desert in Utah (USA), respectively. Both species form package-like structures, often gathered in pseudofilamentous configurations and differ from other Chlorosarcina species by production of ornamented akinetes. Chlorosarcina longispinosa characterized by production of akinetes with long and thin bristles/spines (definition of both; see “Discussion”) and multipartite chloroplast structure, whereas Chlorosarcina brevispinosa typically produced akinetes with shorter bristles/spines and mature vegetative cells, which are possessed by bilobated chloroplast. Both species have parietal chloroplasts, lack pyrenoids and producing zoospores of Protosiphon-type (sensu Starr 1955) without a stigma. Since their original description, both species were only reported few times from arid deserts of different continents (Schwarz 1975; Flechtner et al. 2013).
Deason and Floyd (1987) investigated the zoospore ultrastructure of C. longispinosa and found that the zoospores exhibit typical counterclockwise orientation of the flagellar apparatus and metacentric nuclear division similar to the strain UTEX 1181 Friedmannia israeliensis. The other species of Chlorosarcina (C. stigmatica) according to its ultrastructure is a member of the Chlorophyceae (Deason and Floyd 1987; Sluiman and Blommers 1990). Therefore, Sluiman and Blommers (1990) established a new genus Pleurastrosarcina with its type species P. brevispinosa. They also transferred C. longispinosa to this genus. According to Sluiman and Blommers (1990), this genus was classified to the order Pleurastrales (Ulvophyceae). This order also contained F. israeliensis, which was later transferred to the newly erected class Trebouxiophyceae by Friedl (1995). The genus Pleurastrosarcina escaped the attention of scientists for the long time. Lemieux et al. (2014) demonstrated that Pleurastrosarcina brevispinosa is a member of core Trebouxiophyceae and forms a separate lineage, which they called “Pleurastrosarcina”-clade.
During our study of soil crusts in Las Lomitas at Pan de Azucar National Park and Private Reserve Santa Gracia (Chile), we isolated two strains of Pleurastrosarcina and compared them with available strains of public culture collections using an integrative approach.
Materials and methods
Study area
The samples were collected from two different locations of the Atacama desert (Chile). (1) Las Lomitas, the part of Pan de Azucar National Park is located in southern part of the Atacama desert. The National Park is situated along the Pacific coast and located in the zone of large-scale coastal fog locally called as “Camanchaca”. The landscape of Las Lomitas characterized by steep mountain range on the coastal site reaching up to 850 m asl and low hills going inland, which can reach between 400 and 700 m asl. The typical air humidity varies from 80–85% at night to 60–70% during the day. Annual precipitation is less than 13 mm. The detailed characteristic about climate, water regime, and landscape of the studied site is provided in Lehnert et al. (2018), (2) Private Reserve Santa Gracia is located in the transition zone of semi-arid/mediterranean to arid regions. The landscape is characterized by inland mountain ranges reaching up to 740 m asl with annual precipitation between 70 and 80 mm.
Sampling, isolation, cultivation and morphological observation
The two samples of red and yellow crusts were collected in March 2017. The surface of soil crust around 25 cm2 and 1–2 cm deep was collected in sterile plastic containers. The samples were transported to the lab and were stored by − 25 °C. For establishing of enrichment cultures, 5–7 pieces of 1 cm2 of soil crust were placed in a Petri dish containing Bold Basal Medium (medium 26 in Schlösser 1997) and BG11 (medium 20 in Schlösser 1994). The cultures were incubated at 23 °C, 12:12 h dark:light regime (photon flux rate up 50 μmol m−2 s−1). The cultures were isolated using Pasteur pipette method (Pringsheim 1946). The light microscopical observations of the cultures were done after 2–3 weeks of growth using Olympus BX60 microscope (Olympus, Tokyo, Japan) equipped with digital camera Prog Res C14 plus (Jenoptik, Jena, Germany). Micrographs were taken using the Prog Res Capture Pro imaging system (version 2.9.0.1, Jenoptik, Jena, Germany).
DNA extraction and PCR
The two new isolates were compared with the authentic strains of Pleurastrosarcina longispinosa (UTEX 1183) and P. brevispinosa (UTEX 1176) as well as two strains (SAG 34.83 and ASIB S166), which were isolated by Schwarz (1975) from soil collected Isle of Lavsa, Croatia. The PCR was performed using the MyTaq DNA Polymerase and MyTaq Reaction Buffer (Bioline, Luckenwalde, Germany). The SSU and ITS rDNA were amplified in two overlapping amplicons using green algal-specific primers EAF3 (Marin et al. 2003) and G800R [5′ CATTACTCCGGTCCTACAGACCAACAGG 3′] and G500F [5′ GAATGAGTACAATCTAAACCCCTTAAC 3′] and ITS055R (Marin et al. 2003). PCR products were purified using the MSB® Spin PCRapace (STRATEC Molecular, Berlin, Germany) following the instructions provided by the manufacturer.
Phylogenetic analyses
The new sequences were assembled using the program SeqAssem (Hepperle 2004). The SSU sequences were included in a data set of the Trebouxiophyceae containing representatives of all lineages known for this class. All sequences were aligned according to their secondary structures. The alignment (1747 bp of 58 taxa) was used for phylogenetic analyses. To determinate the evolutionary model that fits best for the data set, the program Modeltest 3.7 (Posada 2008) was used. Considering the results of these tests, the best model was selected by the Akaike information criterion (Akaike 1974). The GTR model with a proportion of invariable sites (I), and gamma shape parameter (G) was used for calculation of the phylogenetic tree. The significance of the presented tree topology was tested using the bootstrap methods by distance [neighbor-joining (NJ) using the GTR + I+G model], parsimony (MP), and maximum likelihood (ML, using the GTR + I+G model). For all calculations, the program PAUP* (version 4.0b164; Swofford 2002) was used. In addition, Bayesian analysis was conducted using the program MrBayes (version 3.2.3; Ronquist et al. 2012).
Along with the SSU rDNA sequences, the complete ITS regions of the six strains were analyzed to detect compensatory base changes (CBCs). The secondary structures of ITS-1 and ITS-2 were folded using the program Mfold (Zuker 2003).
To check if the newly sequenced strains are widely distributed, we analyzed the V4 and V9 regions of the SSU as well as the ITS-2 using the BLAST N search approach (100% coverage, > 97% identity; Altschul et al. 1990).
Results
Molecular phylogeny and species delineation of Pleurastrosarcina
The newly isolated strains (SAG 2586 and SAG 2590) from Chile form together with the authentic strains of P. brevispinosa (UTEX 1176) and P. longispinosa (UTEX 1183) a monophyletic lineage within the Trebouxia lineage of the Trebouxiophyceae. Interestingly, a strain originally assigned as Apatococcus lobatus (SAG 34.83) also belongs to this lineage (Fig. 1). The strain ASIB S166 originally identified as C. brevispinosa forms together with the two Chilean strains the second subclade within the Pleurastrosarcina clade. This lineage is highly supported in all bootstrap and Bayesian analyses. The six strains vary in 15 base positions within their SSU rDNA sequences forming two groups with the Pleurastrosarcina clade called groups I and II. The variable positions within the SSU are mostly located in loop regions of different helices. In addition, the strains belonging to group II differ in one compensatory base change (CBC) in Helix 49 (V9 region; Fig. 2) and one hemi-CBC (one-sided CBC) in the helix E23_13 of the SSU secondary structure. The SSU of strain SAG 34.83 only showed one base difference compared to the strains UTEX 1176 and UTEX 1183, but contains a group I intron at position 516. The strain ASIB S166 also has an intron at the same position. The intron sequences of both strains differ from each other. No introns could be discovered in the other strains.
Molecular phylogeny of the Trebouxiophyceae based on SSU sequence comparisons. The phylogenetic tree shown was inferred by maximum likelihood method based on a data set of 1747 aligned positions of 58 taxa using PAUP 4.0b164. For the analysis, the GTR + I+G model (base frequencies: A 0.2461, C 0.2231, G 0.2758, T 0.2550; rate matrix: A–C 0.9868, A–G 2.1737, A–T 0.8540, C–G 1.4063, C–T 6.6452, G–T 1.0000) with the proportion of invariable sites (I = 0.6288) and gamma distribution shape parameter (G = 0.5597) was chosen, which was calculated as best model by Modeltest 3.7. The Bayesian and bootstrap support was calculated using the same settings. The branches in bold are highly supported (Bayesian values > 0.95; bootstrap values > 70%) in all analyses. The strain designations and accession numbers are given after the species names. The accession numbers in bold represent new sequences of this study, and the newly sequenced strains marked with asterisks contain introns (* = group I intron at position 516)
To obtain a better resolution among the six investigated strains, we analyzed the secondary structures of ITS-1 and ITS-2. As demonstrated in Figs. 3 and 4, the ITS-1 and ITS-2 showed the typical four helix structures among the six strains. The varying regions are highlighted in white boxes in the figures. The comparison of the strains belonging to the two groups showed that the structures of group I strains are more similar to each other than to those of group II. To decide if the strains belong to one or different species, we used the ITS-2/CBC approach, which were described in detail in Darienko et al. (2016). The ITS-2 barcodes presented in Fig. 4 clearly revealed that the groups I and II represent two different species. Both groups differ in two CBCs and two HCBCs from each other. Additional CBCs and HCBCs can be detected in ITS-1 (Fig. 3) and in the variable region of ITS-2 (Fig. 4).
Comparison of the ITS-1 secondary structures among the species of Pleurastrosarcina. The line structure of the ITS-1 has been drawn with PseudoViewer3 (Byun and Han 2009)
Comparison of the ITS-2 secondary structures among the species of Pleurastrosarcina. Extraction of this region and translation into a number code for its usage as barcode. Number code for each base pair: 1 = A–U; 2 = U–A; 3 = G–C; 4 = C–G; 5 = G∙U; 6 = U∙G; 7 = mismatch; 8 = deletion, single or unpaired bases. The line structure of the ITS-2 has been drawn with PseudoViewer3 (Byun and Han 2009). The differences, CBCs, HCBCs, and insertion/deletion (I/D) within the ITS-2 barcodes are marked with an asterisk
To find out if these species are widely distributed, we checked the GenBank entries using the BLAST N search algorithm (100% coverage, > 97% identity). We used for this approach the variable regions V4 and V9 of the SSU rDNA, which are commonly used in environmental studies, and the ITS-2. No additional entries using V4 and ITS-2 could be found. Only three sequences (JX446485-7) using V9 were discovered. We compared the V9 (Helix 49) secondary structure of these sequences with our six investigated strains. As shown in Fig. 2, the structures of these sequences only differ in one base, which represents an HCBC.
Morphological observations
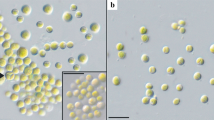
For comparison of the morphology, the six strains of Pleurastrosarcina were cultivated under identical conditions, as described in “Materials and methods”. Figures 5, 6, 7, and 8 demonstrate that all strains (except of UTEX 1183 and ASIB S166) showed similar morphology (three-dimensional cell packages, bi- to trilobate chloroplast without pyrenoid, and without mucilage surrounding the cell packages). The strain UTEX 1183, authentic strain of P. longispinosa, differed by formation of short bristles/spines (2–4 µm long; Fig. 7n–w). The other authentic strain investigated in this study, UTEX 1176 (P. brevispinosa), showed no spine formation, which differs to the original description of Chantanachat and Bold (1962). In contrast, the typical spine formation of P. brevispinosa (Fig. 8c–e) could be discovered in the strain ASIB S166, which was, therefore, identified as this species by Schwarz (1975). Several experiments (incubation for 2 weeks in darkness in distilled water, transfer to different media including marine media, cultivation at lower temperature of 8 °C) to induce the spine formation failed. Interestingly, both Chilean isolates survive full marine media for more than 3 months without any changes in morphology. Zoospore formation could only be discovered by UTEX 1183 and SAG 34.83. Detailed morphological description of each strain is given below in nomenclature and taxonomical consequences of Pleurastrosarcina.
Discussion
Molecular phylogeny and phenotypic plasticity of Pleurastrosarcina
As demonstrated in Fig. 1, the investigated strains formed a highly supported lineage within the Trebouxiophyceae. Our study confirmed the separation of C. brevispinosa and C. longispinosa from other species of Chlorosarcina based on ultrastructural features (Deason and Floyd 1987), which Sluiman and Blommers (1990) have used for the establishment of Pleurastrosarcina. However, Sluiman and Blommers (1990) assigned this genus to the Ulvophyceae, because the Trebouxiophyceae was later described by Friedl (1995). The phylogenetic position of Pleurastrosarcina within the Trebouxiophyceae was already demonstrated by Lemieux et al. (2014) based on chloroplast phylogenomic analyses. The comparison of the SSU and ITS rDNA sequences showed that the six investigated strains belong to two species (groups I and II in Fig. 1). The separation into two species is supported by all phylogenetic analyses and the ITS-2/CBC approach (Figs. 1, 2, 3, 4). In contrast, both species were difficult to identify solely by morphology. Chantanachat and Bold (1962) differentiated both species by differences in spine formation and chloroplast structure (bilobated or lobated). Whereas P. brevispinosa formed short, at the insertion point broad spines, P. longispinosa produced up to 9 µm long, small spines. In contrast as demonstrated in Figs. 5, 6, 7, and 8, only two strains (UTEX 1183 and ASIB S166) showed the typical spine formation. As shown in Fig. 1, both strains belong to different groups indicating that spine formation is a variable feature, which is not suitable for species delineation and not reflected in the phylogenetic analyses. In our morphological comparison, UTEX 1176, the authentic strain of P. brevispinosa, showed no spine formation, in contrast to the original description of Chantanachat and Bold (1962). The spine formation could not be induced by different culture conditions as described above. Despite some morphological differences between UTEX 1176 and UTEX 1183 (see Figs. 6a–m, 7), the SSU and ITS rDNA sequences of both strains are identical.
The formation of spines and bristles is not only variable, and it also has different origins and can be differentiated by staining with Calcofluor White (No. 18909; Sigma-Aldrich, Germany) as demonstrated by Schnepf et al. (1980) and Hegewald and Schnepf (1984, 1987). Hegewald and Schnepf (1984, 1987) defined spines, which were developed before cell wall formation, containing fibrils made from cellulose. In contrast, bristles were formed after the cell wall and did not contain cellulose fibrils. Spines can be stained with Calcofluor White, whereas bristles do not show any staining under fluorescence microscope. With Calcofluor White staining, we could demonstrate that all investigated strains contained cellulose fibrils in their cell wall (data not shown); however, if the ornamentations of cell walls (see Figs. 7n–w, 8c–e) represented spines or bristles, this could not decided because of the low abundance of akinetes in the cultures. It seems that the ornamentations were produced after formation of the cell wall. This indicates that Pleurastrosarcina produces bristles, but this needs further investigations.
The function of spines/bristles in Pleurastrosarcina is unknown. In aquatic habitats, bristle formation can be the response of grazing pressure as demonstrated for Micractinium (Luo et al. 2006; Pröschold et al. 2010). Inducible defense against grazing is not studied on terrestrial green algae so far.
Without spine formation or additional morphological features (ultrastructures of zoospores and cell division), Chlorosarcina-like algae were difficult to identify at generic level. For example, Gärtner and Ingolić (1989) studied the morphology and ultrastructure of several isolates of Apatococcus lobatus, a species usually known as epiphyte of bark of trees and artificial wood constructions such as fences. Only one of their investigated strains was originally isolated from soil (ASIB S183 = SAG 34.83; Schwarz 1975). Despite small differences in morphology, they concluded that the strain SAG 34.83 belongs to Apatococcus lobatus. In contrast, our study clearly revealed that this strain is member of Pleurastrosarcina, which is a separate lineage to Apatococcus, as demonstrated in Fig. 1. This highlights the problematic situation in generic and species delineation based solely on morphology. Green algae-forming cell packages with one chloroplast without pyrenoids are difficult to identify, especially if no further features such as zoospore or spine formation are known. How difficult the identification of taxa with this morphology is, can be demonstrated on the example of Chlorosarcina stigmatica. This species was described by Deason (1959) and the ultrastructure of the type strain (UTEX 962) was investigated by Deason and Floyd (1987). Two other strains were isolated by Schwarz (1975) and Trenkwalder (1975), ASIB S163 and ASIB T105, respectively. Sluiman and Blommers (1990) and Gärtner et al. (1988) demonstrated that the ultrastructure of the cell packages of both ASIB strains showed similarities to UTEX 962; however, no pyrenoid could be observed in both strains in contrast to the findings by type strain of Chlorosarcina stigmatica. Considering the presence of a naked pyrenoid, Deason and Floyd (1987) transferred UTEX 962 to the newly erected genus Desmotetra. Watanabe et al. (2006) demonstrated that UTEX 962 and ASIB T105 belong to Chlorophyceae, but to two different clades and proposed the generic name Sarcinochlamys for strain ASIB T105. Several genera with Chlorosarcina-like morphology were described: Chlorosarcina, Apatococcus, Desmococcus, Diplosphaera, Coccobotrys, and others (see Ettl and Gärtner 2014). Most of these genera are typical terrestrial green algae and belong mostly to the Trebouxiophyceae. However, many of these genera need to be taxonomically revised. As consequence, the usage of an integrative approach as in this study is necessary to assign isolates from different habitats to genera and species.
Ecology and distribution of Pleurastrosarcina
All investigated strains were originally isolated from arid habitats. The authentic strains (P. brevispinosa and P. longispinosa) originated from deserts in Northern America (Chantanachat and Bold 1962), and the two strains isolated by Schwarz (1975) were found in dry soil collected from an island of Croatia. The newly isolated strains from Chilean Atacama desert demonstrated that Pleurastrosarcina seems to be widely distributed, but only in dry soils and arid habitats such as deserts. Flechtner et al. (2013) found three isolates from Colorado desert (California, USA). These strains showed the typical Pleurastrosarcina-like morphology. The comparison of the V9 region of the SSU rDNA confirmed that these strains belong to Pleurastrosarcina (see Fig. 2). Unfortunately, no complete SSU and ITS rDNA sequences are available. Therefore, these isolates cannot be assigned to a species. Summarizing, Pleurastrosarcina is a rare genus and restricted to different types of deserts, but distributed at least on three continents (Europe, North, and South America).
Nomenclature and taxonomical consequences of Pleurastrosarcina
As shown in all figures, the investigated strains form two groups, which represent separate species of the genus Pleurastrosarcina. This genus originally assigned to the Ulvophyceae belongs to the Trebouxiophyceae and form an own lineage. The group I contains the authentic strains of P. brevispinosa and P. longispinosa as well as a strain originally assigned as Apatococcus lobatus. The two isolates from Chile together with strain ASIB S166 originally identified as Chlorosarcina brevispinosa represent the group II. Our new findings require an emendation of the genus Pleurastrosarcina and its type species and a description of P. terriformae as a new species. In addition, we propose several varieties to reflect the morphological variability within the two species:
Pleurastrosarcina Sluiman & Blommers 1990, Arch. Protistenkd. 138: 189.
Emended description: Trebouxiophycean green algae. Cells solitary and spherical compressed in packages. Chloroplasts parietal without pyrenoids. Asexual reproduction by fragmentation of packets or by biflagellated zoospores without cell wall. Zoospores with counterclockwise basal body orientation. Type species: P. brevispinosa (Chantanachat & Bold) Sluiman & Blommers.
Pleurastrosarcina brevispinosa (Chantanachat & Bold) Sluiman & Blommers 1990, Arch. Protistenkd. 138: 189; holotype: NY03049865 = Chicago 1001962 (Fig. 6 a–m).
Basionym: Chlorosarcina brevispinosa Chantanachat & Bold 1962, Univ. Texas Publ. 6218: 40–41.
Emended description: Cells arranged in packages forming pseudofilamentous structures without mucilage. Vegetative cells are oval or spherical, 7.0–9.0 µm in the diameter. Chloroplast bilobated or sometimes trilobated, without pyrenoid. Nucleus is large and good visible. Four-cell packages are 12.0 × 14.0 µm. Reproduction by aplanospore production. Aplanosporangia contain 4–8 spores. Aplanospore are spherical or oval, 4.0–7.0 µm in diameter, containing bilobated chloroplast. Akinetes were not observed. SSU and ITS rDNA sequences (GenBank: MK228837).
Authentic strain: UTEX 1176.
Pleurastrosarcina brevispinosavar.longispinosastat. nov. (Fig. 7 )
Basionym: Chlorosarcina longispinosa Chantanachat & Bold 1962, Univ. Texas Publ. 6218: 42; holotype: NY03049866 = Chicago 1001961.
Synonym: Pleurastrosarcina longispinosa (Chantanachat & Bold) Sluiman & Blommers 1990, Arch. Protistenkd. 138: 189.
Emended description: Cells form sarcinoid-like packages without mucilage. Solitary vegetative cells are 12.0–18.0 µm in diameter. Chloroplast bilobated or cut into several lobes, without the pyrenoid. Nucleus is located in the middle of cell and is good visible. Cytoplasm contains many droplets (probably oil). Cell wall is always thin. Cells form two-to-four-cell packages, which become later three-dimensional structures. In old cultures, cells can reach 25.0 µm in diameter and contain several large vacuoles. The asexual reproduction by zoospore and aplanospore formation. The zoospores and aplanospores are produced in akinetes. Akinetes are spherical or oval, 15.0–18.0 µm in diameter and covered by 3.0–4.0 long spines. Each akinete produce four or eight daughter cells (zoospores or aplanospores). The daughter cells are released by rupture of the mother cell. The remains of akinete cell wall can be observed for the long time in culture. Sometimes, the young cell derived from aplanospores has a rest of the mother cell wall which remains in the top of the cell. The zoospores are of Protosiphon type (without cell wall and become round after a short period of moving), 6.0 × 4.0–9.0 × 4.0 µm in size, have parietal bilobated chloroplasts, anterior nuclei, and are biflagellate, possessing two anterior contractive vacuoles. The young cells are spherical, 7.0–8.0 µm in diameter, with bi- or three-lobated chloroplasts. SSU and ITS rDNA sequences (GenBank: MK228838).
Authentic strain: UTEX 1183.
Pleurastrosarcina brevispinosavar.schwarziivar. nov. (Fig. 5 a–r)
Description: Cells form two- later three-dimensional sarcinoid-like packages. Four-cell packages are 18.0 × 20.0 until 16.0 × 28.0 µm in size. Eight-cell packages can reach 32.0 × 38.0–23.0 × 28.0 µm in size. Cells are spherical if solitary, 11.5–16.0 µm in diameter. Cell wall is relatively thick without ornamentation. Chloroplasts lack pyrenoids and are lobated into two, three, and sometimes more lobes and are pleated. Cell nucleus is large and good visible, often surrounded by small numerous droplets. Asexual reproduction by aplanospores. Aplanosporangia usually contain 4–8 spores, which are released through the rupture of the cell wall. Young cells are broadly ellipsoidal or spherical, 5.0–7.0 µm in diameter, containing usually bilobated chloroplasts. Sometimes, the aplanospores remain in the mother cell and form the packages of the second generation. The liberation of the daughters cells through rupture of the sporangia cell wall. The remains of sporangium cell walls often stick on the surface of the young cells for a long time. Reproduction by zoospore production or sexual reproduction were not observed. SSU and ITS rDNA sequences (GenBank: MK228838).
Authentic strain: SAG 34.83.
Holotype (designated here): The authentic strain SAG 34.83 cryopreserved in a metabolic inactive state at the Culture Collection of Algae (SAG), University of Göttingen, Germany.
Etymology: This variety was named in honor to the isolator of the strain, Dr. Kurt Schwarz.
Type locality: Croatia, Dalmatia, Kornati National Park, Isle Lavsa, soil collected near the coast.
Comment: The three strains of P. brevispinosa vary in their morphology (different sizes and spine formation) and were, therefore, described as different varieties of this species.
Pleurastrosarcina terriformaesp. nov. (Fig. 6 n–t)
Description: Cells form three-dimensional sarcinoid-like packages. Cells are broadly ellipsoidal or spherical if solitary, 12.0–23.0 µm in diameter. Multicellular packages often arranged into short pseudofilamentous structures. Cell wall is usually thin without ornamentation. Chloroplasts lacking a pyrenoid and are lobated into two, three, and sometimes more lobes, pleated. Cell nucleus is large and good visible, often surrounded by small oil droplets. Reproduction by aplanospores. Aplanosporangia usually contain 4–8 spores, which are released through rupture of the cell wall. Young cells are broadly ellipsoidal or spherical, 5.0–7.0 µm in diameter, containing usually bilobated chloroplast. Sometimes, the aplanospores remain in the mother cell and form packages of the second generation, which lead to the formation of three-dimensional structures. The remains of sporangia cell wall often stick on the surface of young cells for the long time. Reproduction by zoospore production or sexual reproduction were not observed. Four-cell packages are usually 20.0 × 22.0 or 18.0 × 23.0 µm in size. The algae stick together in clusters even in 3–4-month-old culture. SSU and ITS rDNA sequences (GenBank: MK228839).
Authentic strain: SAG 2586.
Holotype (designated here): The authentic strain SAG 2586 cryopreserved in a metabolic inactive state at the Culture Collection of Algae (SAG), University of Göttingen, Germany under the barcode number Z000696760.
Etymology: This species was named according to the project EarthShape.
Type locality: Chile, National Park Pan de Azucar, Las Lomitas. The culture was isolated from the red crust on soil.
Pleurastrosarcina terriformaevar.sanctae-graciaevar. nov. (Fig. 5 s–y)
Description: Cells form three-dimensional sarcinoid-like packages. Cells are broadly ellipsoidal or spherical if solitary, 18.0–23.0 µm in diameter. Chloroplasts lobated into two, three, and more lobes. Nucleus is large and located into the middle of cell. Cells contain many small droplets of oil, which provide the culture yellowish color. Reproduction by aplanospores, which are release after the rupture of sporangia cell wall. Other types of asexual or sexual reproduction were not observed. SSU and ITS rDNA sequences (GenBank: MK228840).
Authentic strain: SAG 2590.
Holotype (designated here): The authentic strain SAG 2590 cryopreserved in a metabolic inactive state at the Culture Collection of Algae (SAG), University of Göttingen, Germany under the barcode number Z000694521.
Etymology: This variety was named according to the origin of this strain, Santa Gracia.
Type locality: Chile, National Park Santa Gracia. The culture was isolated from the yellow crust.
Pleurastrosarcina terriformaevar.lavsaevar. nov. (Fig. 8 )
Description: Cell package formation and cell morphology similar to the type variety, but differs by formation of akinetes with spines. SSU and ITS rDNA sequences (GenBank: MK452254).
Authentic strain: ASIB S166.
Holotype (designated here): The authentic strain ASIB S166 cryopreserved in a metabolic inactive state at the Culture Collection of Algae (SAG), University of Göttingen, Germany under the barcode number Z000696412.
Etymology: This variety was named according to the origin of this strain, Isle Lavsa.
Type locality: Croatia, Dalmatia, Kornati National Park, Isle Lavsa, soil collected near the coast.
Comment: The three strains of P. terriformae vary in their morphology (different sizes, spine formation by the variety lavsae) and were, therefore, described as different varieties of this species.
References
Akaike H (1974) A new look at the statistical model identification. IEEE Trans Autom Contr 19:716–723
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Büdel B, Darienko T, Deutschewitz K, Dojani S, Friedl T, Mohr KI, Salisch M, Reisser W, Weber B (2009) Southern African biological soil crust are ubiquitous and highly diverse in dryland, being restricted by rainfall frequency. Microb Ecol 57:229–247
Büdel B, Dulić T, Darienko T, Rybalka N, Friedl T (2016) Cyanobacteria and algae within biological soil crusts. In: Weber B, Büdel B, Belnap J (eds) Ecological Studies. Biological soil crusts: an organizing principle in drylands. Springer, Switzerland, pp 55–80
Byun Y, Han K (2009) PseudoViewer3: generating planar drawings of large-scale RNA structures with pseudoknots. Bioinformatics 25:1435–1437
Chantanachat S, Bold HC (1962) Phycological studies II. Some algae from arid soils. Univ Texas Publ 6218:1–74
Darienko T, Pröschold T (2017) Toward a monograph of non-marine Ulvophyceae using an integrative approach (molecular phylogeny and systematics of terrestrial Ulvophyceae II.). Phytotaxa 324:1–41
Darienko T, Pröschold T (2019) The genus Jaagichlorella Reisigl (Trebouxiophyceae, Chlorophyta) and its close relatives: an evolutionary puzzle. Phytotaxa 388:47–68
Darienko T, Friedl T, Pröschold T (2009) Desmochloris mollenhaueri—a new terrestrial ulvo-phycean alga from south-west African soils. (Molecular phylogeny and systematics of terrestrial Ulvophyceae I.). Algol Stud 129:25–40
Darienko T, Gustavs L, Pröschold T (2016) Species concept and nomenclatural changes within the genera Elliptochloris and Pseudochlorella (Trebouxiophyceae) based on an integrative approach. J Phycol 52:1125–1145
Deason TR (1959) Three Chlorophyceae from Alabama soil. Am J Bot 46:572–578
Deason TR, Floyd GI (1987) Comparative ultrastructure of three species of Chlorosarcina (Chloro- sarcinaceae, Chlorophyta). J Phycol 23:187–195
Ettl H, Gärtner G (2014) Syllabus der Boden-, Luft- und Flechtenalgen. Springer, Heidelberg
Flechtner VR, Pietrasiak N, Lewis LA (2013) Newly revealed diversity of green microalgae from wilderness areas of Joshua Tree National Park (JTNP). Monogr West North Am Naturalist 6:43–63
Friedl T (1995) Inferring taxonomic positions and testing genus level assignments in coccoid green lichen algae: a phylogenetic analysis of 18S ribosomal RNA sequences from Dictyochloris reticulta and from members of the genus Myrmecia (Chlorophyta, Trebouxiophyceae cl. nov.). J Phycol 31:632–639
Fučiková K, Lewis PO, Lewis LA (2014) Widespread desert affiliation of trebouxiophycean algae (Trebouxiophyceae, Chlorophyta) including discovering of three new desert genera. Phycol Res 62:294–305
Gärtner G, Ingolić E (1989) Ein Beitrag zur Kenntnis von Apatococcus lobatus (Chlorophyta, Chaetophorales, Leptosirodeae). Plant Syst Evol 164:133–143
Gärtner G, Hofer A, Ingolić E (1988) Morphological and taxonomical observations on some strains of Chlorosarcina, Chlorosarcinopsis and Planophila (Chlorophyta, Chlorosarcinales), with special reference to “vegetative cell division”. Arch Protistenkd 135:119–131
Groover RD, Bold HC (1969) Phycological Studies VIII. The taxonomy and comparative physiology of the Chlorosarcinales and certain other edaphic algae. Univ Texas Publ 6907:1–165
Hegewald E, Schnepf E (1984) Zur Struktur und Taxonomie bestachelter Chlorellales. Nova Hedwigia 39:297–383
Hegewald E, Schnepf E (1987) Ergänzungen und Korrekturen zur Struktur und Taxonomie bestachelter Chlorellales. Nova Hedwigia 44:537–541
Hepperle D (2004) SeqAssem. A sequence analysis tool, counting assembler and trace data visualization tool for molecular sequences. Win32-version. https://www.sequentix.de
Lehnert LW, Thies B, Trachte K, Achilles S, Osses P, Baumann K, Schmidt J, Samolov E, Jung P, Leinweber P, Karsten U, Büdel B, Bendix J (2018) A case study on fog/low stratus occurrence at Las Lomitas, Atacama Desert (Chile) as a water source for biological soil crusts. Aerosol Air Qual Res 18:254–269
Lemieux C, Otis C, Turmel M (2014) Chloroplast phylogenomic analysis resolves deep-level relationships within the green algal class Trebouxiophyceae. BMC Evol Biol 14:211
Lewis LA, Flechtner VR (2002) Green algae (Chlorophyta) of desert microbiotic crusts: diversity of North American taxa. Taxon 51:443–451
Lewis LA, Lewis PO (2005) Unearthing the molecular phylodiversity of desert soil green algae (Chlorophyta). Syst Biol 54:936–947
Luo W, Pflugmacher S, Pröschold T, Walz N, Krienitz L (2006) Genotype versus phenotype variability in Chlorella and Micractinium (Chlorophyta, Trebouxiophyceae). Protist 157:315–333
Marin B, Palm A, Klingberg M, Melkonian M (2003) Phylogeny and taxonomic revision of plastid-containing euglenophytes based on SSU rDNA sequence comparisons and synapomorphic signatures in the SSU rRNA secondary structure. Protist 154:99–145
Posada D (2008) ModelTest: phylogenetic model averaging. Mol Biol Evol 25:1253–1256
Pringsheim EG (1946) Pure cultures of algae, their preparation and maintenance. Cambridge Univ Press, Cambridge
Pröschold T, Bock C, Luo W, Krienitz L (2010) Polyphyletic distribution of bristle formation in Chlorellaceae: Micractinium, Diacanthos, Didymogenes and Hegewaldia gen. nov. (Trebouxiophyceae, Chlorophyta). Phycol Res 58:1–8
Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542
Schlösser UG (1994) SAG-Sammlung von Algenkulturen at the University of Göttingen, catalogue of strains 1994. Bot Acta 107:113–186
Schlösser UG (1997) Additions to the culture collections of algae since 1994. Bot Acta 110:424–429
Schnepf E, Deichgräber G, Glaab M, Hegewald E (1980) Bristles and spikes in Chlorococcales: ultrastructural studies in Acanthosphaera, Micractinium, Pediastrum, Polyedriopsis, Scene-desmus, and Siderocystopsis. J Ultrastruct Res 72:367–379
Schwarz K (1975) Bodenalgen aus küstennahen Gebieten der Adria. PhD Thesis Phil Fak Univ Innsbruck, Austria
Sluiman HJ, Blommers PCJ (1990) Ultrastructure and taxonomic position of Chlorosarcina stigmatica Deason (Chlorophyceae, Chlorophyta). Arch Protistenkd 138:181–190
Starr RC (1955) A comparative study of Chlorococcum Meneghini and other spherical, zoospore- producing genera of the Chlorococcales. Indiana Univ Publ Sci Ser 20:1–111
Swofford DL (2002) PAUP* phylogenetic analysis using parsimony (*and other methods), version 4.0b10. Sinauer Associates, Sunderland
Trenkwalder H (1975) Untersuchungen zur Bodenalgenflora verschiedener Föhrenwaldtypen im Raum von Brixen (Südtirol, Italien). PhD Thesis Phil Fak Univ Innsbruck, Austria
Watanabe S, Mitsui K, Nakayama T, Inouye I (2006) Phylogenetic relationships and taxonomy of sarcinoid green algae: Chlorosarcinopsis, Desmotetra, Sarcinochlamys gen. nov., Neochlorosarcina, and Chlorosphaeropsis (Chlorophyceae, Chlorophyta). J Phycol 42:679–695
Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acid Res 31:3406–3615
Acknowledgements
Open access funding provided by University of Innsbruck and Medical University of Innsbruck. This work was supported by the Deutsche Forschungsgemeinschaft, (DFG Fr 905/21-1), project 3 within the DFG SPP 1803 (EarthShape: Earth Surface by Biota). We thank N. Kryvenda, D. Nimpsch, and J. C. Reyes-Hagemann for their help by collecting the samples, and T. Friedl for his support of our study.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. Oren.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Darienko, T., Kang, W., Orzechowski, A.K. et al. Pleurastrosarcina terriformae, a new species of a rare desert trebouxiophycean alga discovered by an integrative approach. Extremophiles 23, 573–586 (2019). https://doi.org/10.1007/s00792-019-01108-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-019-01108-5