Abstract
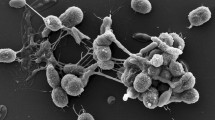
A novel Gram-positive, aerobic, actinobacterial strain, CF5/4T, was isolated in 2007 during an environmental screening of arid desert soil in Ouré Cassoni, Chad. The isolate grew best in a temperature range of 28–40 °C and at pH 6.0–8.5, with 0–1 % (w/v) NaCl, forming brown-coloured and nearly circular colonies on GYM agar. Chemotaxonomic and molecular characteristics of the isolate matched those described for members of the genus Geodermatophilus. The DNA G + C content of the novel strain was 75.9 mol %. The peptidoglycan contained meso-diaminopimelic acid as diagnostic diaminoacid. The main phospholipids were phosphatidylethanolamine, phosphatidylcholine, phosphatidylinositol, diphosphatidylglycerol and a small amount of phosphatidylglycerol; MK-9(H4) was identified as the dominant menaquinone and galactose as diagnostic sugar. The major cellular fatty acids were branched-chain saturated acids: iso-C15:0 and iso-C16:0. The 16S rRNA gene showed 96.2–98.3 % sequence identity with the three members of the genus Geodermatophilus: G. obscurus (96.2 %), G. ruber (96.5 %), and G. nigrescens (98.3 %). Based on the chemotaxonomic results, 16S rRNA gene sequence analysis and DNA–DNA hybridization with the type strain of G. nigrescens, the isolate is proposed to represent a novel species, Geodermatophilus arenarius (type strain CF5/4T = DSM 45418T = MTCC 11413T = CCUG 62763T).


Similar content being viewed by others
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Castresana J (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17:540–552
Collins MD, Pirouz T, Goodfellow M, Minnikin DE (1977) Distribution of menaquinones in actinomycetes and corynebacteria. J Gen Microbiol 100:221–230
De Ley J, Cattoir H, Reynaerts A (1970) The quantitative measurement of DNA hybridization from renaturation rates. Eur J Biochem 12:133–142
DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL (2006) Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072
Giongo A, Favet J, Lapanje A, Gano KA, Kennedy S, Davis-Richardson AG, Brown C, Beck A, Farmerie WG, Cattaneo A, Crabb DB, Aung YY, Kort R, Brumsack HJ, Schnetger B, Broughton WJ, Gorbushina AA, Triplett EW (2012) Microbial hitchhikers on intercontinental dust: high- throughput sequencing to catalogue microbes in small sand samples. Aerobiologia. doi:10.1007/s10453-012-9264-0. Accepted June 6, 2012
Gordon RE, Smith MM (1955) Proposed group of characters for the separation of Streptomyces and Nocardia. J Bacteriol 69:147–150
Gregersen T (1978) Rapid method for distinction of gram-negative from positive bacteria. Appl Microbiol Biotechnol 5:123–127
Hess PN, De Moraes Russo CA (2007) An empirical test of the midpoint rooting method. Biol J Linn Soc 92:669–674
Huss VAR, Fest H, Schleifer KH (1983) Studies on the spectrophotometric determination of DNA hybridization from renaturation rates. Syst Appl Microbiol 4:184–192
Ivanova N, Sikorski J, Jando M, Munk C, Lapidus A, Glavina Del Rio T, Copeland A, Tice H, Cheng JF, Lucas S et al (2010) Complete genome sequence of Geodermatophilus obscurus type strain (G-20T). Stand Genomic Sci 2:158–167
Kroppenstedt RM (1982) Separation of bacterial menaquinones by HPLC using reverse phase (RP18) and a silver loaded ion exchanger. J Liq Chromatogr 5:2359–2387
Kroppenstedt RM, Goodfellow M (2006) The family Thermomonosporaceae: Actinocorallia, Actinomadura, Spirillispora and Thermomonospora. In: Dworkin M, Falkow S, Schleifer KH, Stackebrandt E (eds) The prokaryotes, 3rd edn, vol 3. Archaea and Bacteria: Firmicutes, Actinomycetes. Springer, New York, pp 682–724
Lechevalier MP, Lechevalier HA (1970) Chemical composition as a criterion in the classification of aerobic actinomycetes. Int J Syst Bacteriol 20:435–443
Lee C, Grasso C, Sharlow MF (2002) Multiple sequence alignment using partial order graphs. Bioinformatics 18:452–464
List Editor (2012) List of new names and new combinations previously effectively, but not validly, published. Int J Syst Evol Microbiol 62:2045–2047
Luedemann GM (1968) Geodermatophilus, a new genus of the Dermatophilaceae (Actinomycetales). J Bacteriol 96:1848–1858
Mesbah M, Premachandran U, Whitman WB (1989) Precise measurement of the G + C content of deoxyribonucleic acid by high-performance liquid chromatography. Int J Syst Bacteriol 39:159–167
Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G, Athalye M, Schaal K, Parlett JH (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241
Nie GX, Ming H, Li S, Zhou EM, Cheng J, Yu TT, Zhang J, Feng HG, Tang SK, Li WJ (2012) Geodermatophilus nigrescens sp. nov., isolated from a dry-hot valley. Antonie Van Leeuwenhoek 101:811–817
Normand P (2006) Geodermatophilaceae fam. nov., a formal description. Int J Syst Evol Microbiol 56:2277–2278
Normand P, Orso S, Cournoyer B, Jeannin P, Chapelon C, Dawson J, Evtushenko L, Misra AK (1996) Molecular phylogeny of the genus Frankia and related genera and emendation of the family Frankiaceae. Int J Syst Bacteriol 46:1–9
Pattengale ND, Alipour M, Bininda-Emonds ORP, Moret BME, Stamatakis A (2009) How many bootstrap replicates are necessary? Lect Notes Comput Sci 5541:184–200
Pelczar MJ Jr (ed) (1957) Manual of microbiological methods. McGraw-Hill Book Co., New York
Rainey FA, Ward-Rainey N, Kroppenstedt RM, Stackebrandt E (1996) The genus Nocardiopsis represents a phylogenetically coherent taxon and a distinct actinomycete lineage: proposal of Nocardiopsiaceae fam. nov. Int J Syst Bacteriol 46:28–96
Rice P, Longden I, Bleasby A (2000) EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16:276–277
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. USFCC Newsl 20:16
Schleifer KH, Kandler O (1972) Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev 36:407–477
Shirling EB, Gottlieb D (1966) Methods for characterization of Streptomyces species. Int J Syst Bacteriol 16:313–340
Stamatakis A, Hoover P, Rougemont J (2008) A rapid bootstrap algorithm for the RAxML web servers. Syst Biol 57:758–771
Staneck JL, Roberts GD (1974) Simplified approach to identification of aerobic actinomycetes by thin-layer chromatography. Appl Microbiol 28:226–231
Swofford DL (2002) PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4.0 b10. Sinauer Associates, Sunderland
Tindall BJ, Sikorski J, Simbert RA, Krieg NR (2007) Phenotypic characterization and the principles of comparative systematics. In: Beveridge TJ, Breznak JA, Marzluf GA, Schmidt TM, Snyder LR (eds) Reddy CA (Editor in Chief) Methods for general and molecular microbiology. ASM Press, Washington, pp 330–393
Urzì C, Brusetti L, Salamone P, Sorlini C, Stackebrandt E, Daffonchio D (2001) Biodiversity of Geodermatophilaceae isolated from stones and monuments in the Mediterranean basin. Environ Microbiol 3:471–479
Vaas LAI, Sikorski J, Michael V, Göker M, Klenk HP (2012) Visualization and curve-parameter estimation strategies for efficient exploration of phenotype microarray kinetics. PLoS ONE 7:e34846
Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, Moore LH, Moore WEC, Murray RGE, Stackebrandt E, Starr MP, Trüper HG (1987) Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol 37:463–464
Zhang YQ, Chen J, Liu HY, Zhang YQ, Li WJ, Yu LY (2011) Geodermatophilus ruber sp. nov., isolated from rhizosphere soil of a medical plant. Int J Syst Evol Microbiol 61:190–193
Acknowledgments
We would like to gratefully acknowledge the help of Agathe Stricker of the International Committee of the Red Cross in Geneva, Switzerland, for organizing the collection of the sand samples from which strain CF5/4T was isolated. Jocelyne Favet, Arlette Cattaneo and William J. Broughton of the Laboratoire de Biologie Moléculaire de Plantes Supérieures (University of Geneva, Switzerland) are acknowledged for their contribution to the isolation of the strain, and Brian J. Tindall (DSMZ, Braunschweig) for his guidance in chemotaxonomical analyses. M.C. Montero-Calasanz is the recipient of a postdoctoral contract from European Social Fund Operational Programme (2007–2013) for Andalusia.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Communicated by A. Oren.
The INSDC accession number for the 16S rRNA gene sequence of strain CF5/4T is HE654547.
Electronic supplementary material
Below is the link to the electronic supplementary material.
792_2012_486_MOESM1_ESM.pdf
Fig. S1. The parameter “Maximum Height” estimated from the respiration curves as measured with the OmniLog phenotyping device and visualized as heatmap using the opm package. Plates and substrates are rearranged according to their overall similarity (as depicted using the row and column dendrograms). First picture, heatmap inferred from the discretized values; second picture, heatmap inferred from the original “Maximum Height” measurements, standardized per row; third picture, heatmap inferred from the original “Maximum Height” measurements, not standardized. (PDF 36 kb)
792_2012_486_MOESM2_ESM.tif
Fig. S2. Polar lipids profile of Geodermatophilus arenarius sp. nov. CF5/4T after separation by two-dimensional TLC. Plate was sprayed with molybdatophosphoric acid for detection of total polar lipids. DPG, diphosphatidylglycerol; PE, phosphatidylethanolamine; PC, phosphatidylcholine; PI, phosphatidylinositol; PG, phosphatidylglycerol; GL, glycolipid. (TIFF 594 kb)
792_2012_486_MOESM3_ESM.docx
Table S1. Fatty acids profiles (%) of strain CF5/4T and the type strains of other Geodermatophilus species. Strains: 1, G. arenarius sp. nov. CF5/4T (DSM 45418T); 2, G. obscurus G-20T (DSM 43160T); 3, G. ruber CPCC 21356T (DSM 45317T); 4, G. nigrescens YIM 75980T (DSM 45408T). (DOCX 20 kb)
Rights and permissions
About this article
Cite this article
Montero-Calasanz, M.C., Göker, M., Pötter, G. et al. Geodermatophilus arenarius sp. nov., a xerophilic actinomycete isolated from Saharan desert sand in Chad. Extremophiles 16, 903–909 (2012). https://doi.org/10.1007/s00792-012-0486-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-012-0486-4