Abstract
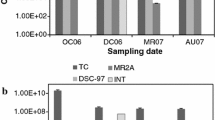
We have studied the diversity of culturable halophilic Archaea at Rambla Salada, Murcia (south-eastern Spain). We made 8 samplings at different places in this habitat during the years 2006 and 2007 and isolated a total of 49 strains, which were identified by means of phenotypic tests and the hypervariable V1–V3 region of the 16S rRNA gene sequences (around 500 bp). The ribosomal data showed that the isolates belonged to 12 genera within the Halobacteriaceae family, with Haloferax and Natrinema being the most abundant. Five strains showed less than 97% sequence identity with validly described species and may well represent new taxa. All the strains grew best with around 25% w/v salts, required high concentrations of NaCl and magnesium and produced red to pink colonies. They were facultative anaerobes with both respiratory and fermentative metabolisms. The diversity of the archaeal community was analysed with the MOTHUR package. We identified 14 OTUs at the 3% genetic distance level and found quite high diversity. Rarefaction curves of richness estimators and diversity indices demonstrated that our collection of isolates represented the archaeal community at Rambla Salada that can be isolated under the conditions used in this work. This is the first report to be published on the culturable archaea at Rambla Salada, an area of considerable ecological interest.



Similar content being viewed by others
References
Aller JY, Kemp PF (2008) Are Archaea inherently less diverse than Bacteria in the same environments? FEMS Microbiol Ecol 65:74–87
Antón J, Oren A, Benlloch S, Rodríguez-Valera F, Amann R, Rossselló-Mora R (2002) Salinibacter ruber gen. nov., sp. nov., a novel, extremely halophilic member of the Bacteria from saltern crystallizer ponds. Int J Syst Evol Microbiol 52:485–491
Auguet JC, Barberan A, Casamayor EO (2010) Global ecological patterns in uncultured Archaea. ISME J 4:182–190
Baati H, Guermazi S, Amdouni R, Gharsallah N, Sghir A, Ammar E (2008) Prokaryotic diversity of a Tunisian multipond solar saltern. Extremophiles 12:505–518
Baati H, Guermazi S, Gharsallah N, Sghir A, Ammar E (2010) Novel prokaryotic diversity in sediments of Tunisian multipond solar saltern. Res Microbiol 161:573–582
Burns DG, Camakaris HM, Janssen PH, Dyall-Smith ML (2004) Combined use of cultivation-dependent and cultivation-independent methods indicates that members of most haloarchaeal groups in an Australian crystallizer pond are culturable. Appl Environ Microbiol 70:5258–5265
Cambon-Bonavita MA, Nadalig T, Roussel E, Delage E, Duperron S, Caprais JC, Boetius A, Sibuet M (2009) Diversity and distribution of methane-oxidizing microbial communities associated with different faunal assemblages in a giant pockmark of the Gabon continental margin. Deep sea res part II: topical studies in oceanography 56:2248–2258
Chao A (1987) Estimating the population size for capture–recapture data with unequal catchability. Biometrics 43:783–791
Chun J, Lee JH, Jung Y, Kim M, Kim S, Kim BK, Lim YW (2007) EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int J Syst Evol Microbiol 57:2259–2261
Clementino MM, Vieira RP, Cardoso AM, Nascimento AP, Silveira CB, Riva TC, González AS, Paranhos R, Albano RM, Ventosa A, Martins OB (2008) Prokaryotic diversity in one of the largest hypersaline coastal lagoons in the world. Extremophiles 12:595–604
Dave SR, Desai HB (2006) Microbial diversity at marine saltern near Bhavnagar, Gujarat, India. Curr Sci 90:497–500
González-Domenech CM, Martínez-Checa F, Quesada E, Béjar V (2008) Halomonas cerina sp. nov., a moderately halophilic, denitrifying, exopolysaccharide-producing bacterium. Int J Syst Evol Microbiol 58:803–809
Grant S, Grant WD, Jones BE, Kato C, Li L (1999) Novel archaeal phylotypes from an East African alkaline saltern. Extremophiles 3:139–145
Grant WD, Kamekura M, McGenity TJ, Ventosa A (2001) Order I. Halobacteriales. In: Garrity GM (ed) Bergey’s manual of systematic bacteriology V.I. The Archaea and deeply branching and phototrophic bacteria. Springer, New York, pp 213–235
Hughes JB, Hellmann JJ, Ricketts TH, Bohannan BJ (2001) Counting the uncountable: statistical approaches to estimating microbial diversity. Appl Environ Microbiol 67:4399–4406
Ihara K, Watanabe S, Tamura T (1997) Haloarcula argentinensis sp. nov. and Haloarcula mukohataei sp. nov., two new extremely halophilic archaea collected in Argentina. Int J Syst Bacteriol 47:73–77
Litchfield CD, Irby A, Vreeland RH (1999) The microbial ecology of solar salt plants. In: Oren A (ed) Microbiology and biogeochemistry of hypersaline environments. CRC Press, Boca Ratón, pp 39–52
Lozupone CA, Knight R (2007) Global patterns in bacterial diversity. Proc Natl Acad Sci USA 104:11436–11440
Magurran AE (1996) Ecological diversity and its measurement. Chapman and Hall, London
Marmur J (1961) A procedure for the isolation of deoxyribonucleic acid from microorganisms. J Mol Biol 3:207–208
Martínez-Cánovas MJ, Béjar V, Martínez-Checa F, Páez R, Quesada E (2004) Idiomarina fontislapidosi sp. nov. and Idiomarina ramblicola sp. nov., isolated from inland hypersaline habitats in Spain. Int J Syst Evol Microbiol 54:1793–1797
Martín-Platero AM, Valdivia E, Maqueda M, Martínez-Bueno M (2007) Fast, convenient, and economical method for isolating genomic DNA from lactic acid bacteria using a modification of the protein “salting-out” procedure. Anal Biochem 366:102–104
Maturrano L, Santos F, Rosselló-Mora R, Antón J (2006) Microbial diversity in Maras salters, a hypersaline environment in the Peruvian Andes. Appl Environ Microbiol 72:13887–13895
Moraine RA, Rogovin P (1966) Kinetics of polysaccharide B-1459 fermentation. Biotechnol Bioeng 8:511–524
Muller DW, Hsü KJ (1987) Event stratigraphy and paleoceanography in the Fortuna basin (Southeast Spain): a scenario for the Messinian salinity crisis. Paleoceanography 2:679–696
Oren A (1994) The ecology of the extremely halophilic archaea. FEMS Microbiol Rev 13:415–440
Oren A (2002) Halophilic microorganisms and their environments. Kluwer Academic Publishers, Netherlands
Oren AT (2006) The order Halobacteriales. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E (eds) The prokaryotes a handbook on the biology of bacteria. Springer, New York, pp 113–164
Oren A (2011) Diversity of halophiles. In: Horikoshi K (ed) Extremophiles handbook. Springer, Tokyo, pp 309–325
Oren A, Ventosa A, Grant WD (1997) Proposed minimal standards for description of new taxa in the order Halobacteriales. Int J Syst Evol Microbiol 47:233–238
Oren A, Arahal DR, Ventosa A (2009) Emended descriptions of genera of the family Halobacteriaceae. Int J Syst Evol Microbiol 59:637–642
Ozcan B, Ozcengiz G, Coleri A, Cokmus C (2007) Diversity of halophilic archaea from six hypersaline environments in Turkey. J Microbiol Biotechnol 17:985–992
Pasić L, Bartual SG, Ulrih NP, Grabnar M, Velikonja BH (2005) Diversity of halophilic archaea in the crystallizers of an Adriatic solar saltern. FEMS Microbiol Ecol 54:491–498
Rodríguez-Valera F, Ventosa A, Juez G, Imhoff J (1985) Variation of the environmental features and microbial population with salt concentrations in a multipond saltern. Microbiol Ecol 11:107–115
Rodríguez-Varela F, Ruiz-Berraquero F, Ramos-Cormenzana A (1981) Characteristics of the heterotrophic bacterial populations in hypersaline environments of different salt concentration. Microbiol Ecol 7:235–243
Schloss PD, Handelsman J (2005) Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol 71:1501–1506
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing MOTHUR: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Simpson EH (1949) Measurement of diversity. Nature 163:688
Tamames J, Abellán JJ, Pignatelli M, Camacho A, Moya A (2010) Environmental distribution of prokaryotic taxa. BMC Microbiol 10:85
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Velasco J, Millán A, Hernández J, Gutiérrez C, Abellán P, Sánchez D, Ruíz M (2006) Response of biotic communities to salinity changes in a Mediterranean hypersaline stream. Saline Syst 2:12
Acknowledgments
The authors thank Josefa Velasco and Andrés Millán for their assistance with field work and zone selection for sampling. Our thanks also go to Miguel Angel Núñez of the Explanation Centre at Rambla Salada for his willing assistance in the field. This research was supported by grants from the Dirección General de Investigación Científica y Técnica (CGL2005-05947; CGL2008-02399), from the Plan Andaluz de Investigación and Plan Propio de la Universidad de Granada, Spain. We also thank our colleague Dr J. Trout for revising our English text.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. Oren.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Luque, R., González-Domenech, C.M., Llamas, I. et al. Diversity of culturable halophilic archaea isolated from Rambla Salada, Murcia (Spain). Extremophiles 16, 205–213 (2012). https://doi.org/10.1007/s00792-011-0420-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-011-0420-1