Abstract
Objective
Aim of this study was to investigate the impact of human PDL-derived fibroblasts (HPDF) and human alveolar bone-derived osteoblasts (HABO) co-culture on the expression of cytokines involved in tissue remodeling using an in vitro compressive force (CF) model.
Materials and methods
Static compressive force (CF) of 47.4 g/cm2 was applied on mono- and co-cultured HPDFs and HABOs for 1, 2, or 4 h at 30 °C. TNFA, PTGS2, and IL6 gene expressions were determined by quantitative real-time polymerase chain reaction. TNF, PGE2, and IL6 concentrations were measured using enzyme-linked immunosorbent assay.
Results
In mono-culture, TNFA, PTGS2, and IL6 gene expressions were upregulated under CF as compared to controls for each time period in both cell types. PGE2 increased at 1 and 2 h in both cell types, and IL6 increased only at 2 and 4 h in HPDFs. Co-culture alleviated the force-induced increase of the expression of TNFA, PTGS2, IL6, PGE2, and IL6 in HPDFs at any time point. In HABOs, co-cultivation decreased the expression of PGE2 after 1 h and 4 h, and that of IL6 after 1 h compared to mono-culture.
Conclusions
CF application on co-cultures of HPDFs and HABOs causes significant changes of TNFA, PTGS2, and IL6 gene expressions and PGE2 and IL6 production in comparison to mono-culture indicating intercellular communication.
Clinical relevance
Mechanical stimulation of HPDFs and HABOs in co-culture induces a different gene expression pattern than stimulation of individual cell types alone. Co-culture might therefore be a relevant method to elucidate periodontal regeneration during orthodontic therapy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The periodontal supporting tissue including the alveolar bone keeps the teeth in their position in order to maintain their normal physiological function [1]. Teeth are surrounded by a layer of periodontal ligament, consisting of a heterogeneous population of different cell types, among which are periodontal ligament fibroblasts together with blood vessels, nerves, and connective tissue fibers [2]. During orthodontic therapy, mechanical forces are applied to the teeth. Within certain limits, the periodontal ligament absorbs and distributes these forces to the surrounding tissue compartments and induces a subtle inflammatory response which is essentially mediated by various cytokines and enzymes ultimately leading to tissue remodeling in the surrounding alveolar bone [3, 4].
Numerous studies [5,6,7,8] evidenced that particularly pro-inflammatory cytokines and enzymes normally involved in physiological tissue homeostasis play pivotal roles in orthodontically induced bone remodeling. Among others, the cytokine tumor necrosis factor (TNF) is not only present under inflammatory conditions [9] but plays a central role also during the exposure of the periodontal ligament against orthodontic force [8]. Detailed analyses of the role of TNF in this context shows that it gathers apoptotic bone cells under mechanical pressure, inhibits osteoblasts, and stimulates osteoclastogenesis [10], ultimately leading to a net bone resorption. Also, interleukin 6 (IL6) is linked to the homeostasis of osseous tissue [11, 12]. The lack of IL6 can impair the bone mass via reduction of the number of osteoblasts [13, 14]. Moreover, a previous study identified a significant role of IL6 in the mediation of periodontal tissue remodeling during orthodontic tooth movement [6]. Cyclooxygenase-2 (prostaglandin-endoperoxide synthase 2, PTGS2) is one of the key regulators for the production of the various prostaglandin entities including prostaglandin E2, which is a strong inducer of bone resorbing processes [7, 15]. PTGS2 is ubiquitously expressed and is present within the periodontal tissue. Apart from genuine inflammatory reactions, its expression is also amplified under the influence of mechanical force [7, 15] ultimately leading to an increasing production of PGE2 under mechanical loading [16].
The metabolic pathways of periodontal ligament fibroblasts, osteoblasts, and osteoclasts are cross-linked via various signaling cascades controlling and maintaining their physiology [17, 18]. So far, intercellular communication during mechanical force loading cannot be fully elucidated. Most in vitro studies use a single cell type (mono-culture) to investigate the effect of mechanical stimulation. Recently, communication between cells has received increasing attention using co-culture techniques [18, 19]. Two kinds of experimental co-cultivation setups are available to explore the reciprocities of two cell types under loading conditions: direct- and indirect-contact co-culture. Direct-contact co-culture (two cell types are grown in contact to each other) is commonly used to investigate the molecular signaling pathways between cells [18]. Nevertheless, it is technically demanding to separate different cell types which have been grown together in a direct-contact co-culture following force application. Indirect-contact co-culture [20] circumvents this obstacle since two different types of cells share the same growth environment having no direct contact to each other. Cells share the same culture medium, but cytokines, growth factors, and other mediator molecules can pass through a porous bio-membrane, separating both cell types. This particular method of co-cultivation allows analyzing and assessing different cells separately [20].
This study aimed to establish an indirect-contact co-culture system of human PDL-derived fibroblasts (HPDFs) and alveolar bone-derived osteoblasts (HABOs) under static compressive force (CF) application. Using this, it should be tested if intercellular communication between HPDFs and HABOs exists and if it has an impact on the regulation of various mechanically induced cytokines, enzymes, and metabolites, i.e., TNF, PTGS2, IL6, and PGE2. The null hypothesis states that there is no difference in expression and activation of these mediators between cells in mono-culture and co-culture, respectively. The primary endpoint was that the co-culture of both cell types has significant impact on the expression of the marker molecules tested herein. The null hypothesis was that there is no difference when culturing the cells alone or together. The secondary endpoint can be defined as the influence of the compressive force on the expression of the tested marker molecules. The null hypothesis was that the compressive force has no influence on the expression of these molecules. A third endpoint was formulated regarding the effect of the compressive force on the apoptosis. The null hypothesis was that the force has no impact on the rate of cellular apoptosis.
Materials and methods
Primary cell culture
Human periodontal ligament-derived fibroblasts (HPDFs) were derived from first molars obtained from healthy periodontal sites of patients extracted for orthodontic reasons. They were generated using the explant technique according to Somerman et al. [21]. Human alveolar bone-derived osteoblasts (HABOs) derived from alveolar bone obtained from a patient undergoing orthognathic surgery [22]. The protocol was approved by the Ethics Committee of the University Hospital of the Ludwig-Maximilians-Universität Munich (no. 045-09). Prior to enrolment into the study, each study subject gave written informed consent. HPDFs were cultivated in Dulbecco’s Modified Eagle’s medium (Biochrom, Berlin, Germany) supplemented with 10% fetal bovine serum (FBS) (Sigma-Aldrich, St. Louis, MO, USA) and 2% l-glutamine (Sigma-Aldrich, St. Louis, MO, USA). HABOs were cultivated in Dulbecco’s Modified Eagle’s Medium/Nutrient Mixture F-12 Ham (DMEM/F-12) (Sigma-Aldrich, St. Louis, MO, USA) with 10% FBS and 2% MEM-vitamins (Biochrom, Berlin, Germany). Both cell types were grown in a standard cell culture incubator with 5% CO2 at 37 °C in a humidified atmosphere. Cells were passaged at regular intervals depending on their growth characteristics using 0.05% trypsin-EDTA solution (Biochrom, Berlin, Germany). Cells at the fifth passage were used in all experiments.
Force application with the centrifugal model
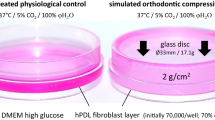
To mimic orthodontic force application, a modified centrifugal model for induction of compressive force (CF) onto the cell culture was used [23,24,25] (Fig. 1).
Schematic drawing of the experimental setup for force application through centrifugation [modified after 34]. The right panel shows the situation for the mono-culture (top) and co-culture (bottom) setup. During centrifugation, the buckets swing out due to the centrifugal force \( \overrightarrow{F} \) that is perpendicular to the growth plane of the cells in culture. According to the manufacturer, the distance between insert and the bottom of the well is less than 1 mm (Greiner Bio-One)
Temperature control and stability during centrifugation
In pre-tests, the temperature setting of the centrifuge (Sigma 4-16 K, Sigma, Osterode am Harz, Germany) was calibrated using a temperature data logger (Thermochron iButton®, DS1921H-F5, Maxim Integrated, San Jose, USA). This device measures the temperature at pre-defined intervals (here set to 1 min) with an accuracy of ± 1 °C. Due to technical reasons, the maximum temperature that could be kept stable for up to 8 h was 30 °C at 200×g. Accordingly, all cell culture experiments were done at 30 °C. Negative control samples were cultured at 30 °C using an incubator (Binder, Tuttlingen, Germany). The temperature within the incubator was continuously monitored.
Preparation of mono-cultured cells for CF application
HPDFs and HABOs were seeded separately in 12-well plates at a density of 1 × 105 cells/well. After 6 h, the medium was changed to medium with reduced FBS (1%) and cultivation was continued for 10 h. Afterwards, they were subjected to CF application or served as negative controls as described above.
Preparation of cells in co-culture for CF application
HPDFs were seeded in 12-well plates at a density of 1 × 105 cells/well, and HABOs at 5 × 104 cells/insert were seeded in ThinCert™ cell culture inserts (pore size, 1.0 μm; culture area, 113.1 mm2; Greiner Bio-One, Frickenhausen, Germany), thus preventing cross-membrane migration of cells. After attachment (about 6 h), cell culture media were removed. DMEM/F-12 containing 1% FBS was added to the wells. The seeded inserts were placed into the corresponding wells and DMEM/F-12 medium was added to each well giving a total volume of 900 μL/well. They were co-cultivated for at least 10 h before centrifugation. Centrifugation was done as described above. The co-cultures were repeated with switched positions (HPDFs in inserts and HABOs in wells) using identical cell densities for insert and well as given above. Cell culture supernatant of the whole well and cell lysates for total RNA preparations were extracted from the cells in the wells after removal of the inserts.
CF application
All centrifugations were done at 200×g for 1, 2, and 4 h. This setting equals to 47.4 g/cm2 CF [25]. To shorten total centrifugation time and to reach the temperature of 30 °C more quickly, the centrifuge was pre-run at 800×g for 47 min prior to the placement of cells. Cells in the negative control groups received similar processing and were kept in the incubator at 30 °C for the same time periods. For each cell type/culture combination six replicates were done.
Apoptosis
Cellular apoptosis induction was tested after incubation at 30 °C for up to 4 h in comparison to 37 °C. HPDFs and HABOs were seeded in 12-well plates with 1 × 105 cells/well, followed by an overnight incubation at 37 °C with 5% CO2. Plates representing the experimental (30 °C) and negative control group (37 °C with 5% CO2) were incubated for 4 h at the given temperatures. An apoptosis-positive control was established using stimulation with 4 mM dithiothreitol (Applichem, Darmstadt, Germany) for 24 h at 37 °C and 5% CO2 in a humidified atmosphere [26]. Cells were stained using the Multi-Parameter Apoptosis Assay Kit (Cayman, Ann Arbor, MI, USA) according to the manufacturer’s protocol following the single-step staining protocol. Nuclear fragmentation was detected by Hoechst dye staining, reversal of the mitochondrial membrane potential with TMRE, and the flipping of the membrane lipids using Annexin V-FITC. Immediately after staining, the cells were examined using an EVOSfl fluorescence microscope (Invitrogen, Carlsbad, CA, USA) at × 200 and × 400 magnifications. All test procedures were repeated at least three times for each cell types.
Real-time polymerase chain reaction
Total RNA lysates of HPDFs and HABOs were prepared using the Quick-RNA™ MicroPrep Kit (Zymo, Irvine, CA, USA). After removal of the supernatant (and inserts, if applicable), the cells seeded on the bottom of the wells were lysed using the lysing solution provided with the Quick-RNA kit (Zymo, Irvine, CA, USA). The cell lysates were passed through QIAshredder™ columns (Qiagen, Hilden, Germany). To remove impurities by genomic DNA, DNase I digestion was done as suggested by the manufacturer (Zymo, Irvine, CA, USA).
Total RNA was quantified using the NanoDrop1000 (Peqlab, Erlangen, Germany). From each sample, 600 ng total RNA was reverse-transcribed into complementary DNA (cDNA) using the SuperScript IV First-Strand Synthesis System (Thermo Fisher Scientific, Waltham, MA, USA). The cDNA was then diluted 1:5 for real-time polymerase chain reaction (real-time PCR) amplification.
Real-time PCR was carried out to determine the relative expression of the TNFA, PTGS2, and IL6 genes in the cell samples using Luminaris Color HiGreen qPCR-Mastermix (Thermo Fisher Scientific, Waltham, MA, USA) in a LightCycler® 480 (Roche Molecular Diagnostics, Basel, Switzerland) according to the manufacturer’s protocol with the following modifications: in addition to the qPCR mastermix, each PCR reaction contained 2 μL of diluted cDNA and 6 μM of each forward and reverse primers (Table 1). A four-step cycling protocol was used that included an additional data acquisition step (5 s) after the extension step 5 °C below the melting point of the specific PCR product (Table 1) in a total of 45 PCR cycles. From a panel of ten different reference genes (HHK-1, Real-Time Primers, LLC, Elkins Park, PA, USA), beta-2-microglobulin (B2M) showed no change in gene expression between experimental condition and negative controls at all and was therefore used as the reference gene. The ΔΔCq method was applied for quantification of gene expression [18]. To evaluate the primer efficiencies, standard curves prepared from serial dilutions of cDNA (undiluted, 1:10, 1:100, 1:1000, and 1:10000) were quantified in the LightCycler® 480 (Table 1). All cDNA samples were measured twice, giving a total of 12 PCR amplifications for each gene-cell type-culture combination.
Enzyme-linked immunosorbent assay
Cell culture supernatants were collected for enzyme-linked immunosorbent assay (ELISA) quantification. The concentration of IL6, TNF, and PGE2 was measured using Duo Set ELISA Kits (IL6, TNFα) or the PGE2 Parameter Assay kit according to manufacturer’s instructions (R&D Systems, Minneapolis, MN, USA). Measurements were done using the Varioscan microplate reader (Thermo Electron Corporation, Vantaa, Finland). All samples were measured in triplicate. In co-culture setups, the cell type reported is the one seeded on the bottom of the wells.
Statistics
Results were presented as mean ± standard error of mean (SEM). To evaluate significant changes between groups (CF vs. control; mono-culture vs. co-culture), the independent-samples Mann-Whitney U test was applied using IBM SPSS Statistics 25 (IBM Corp., Armonk, NY, USA). All test procedures were two-tailed considering p values < 0.05 significant. Since three genetic loci have been tested herein, the p values were corrected for multiple testing applying the Bonferroni procedure finally adjusting the level of significance pcorr to 0.017.
Results
Determination of temperature
Temperature development was recorded throughout all experiments (Fig. 2). The room temperature was kept constant at 23 (± 1) °C. During application of compressive force (CF), the temperature inside the centrifuge running at 200×g was 30.5 (± 1) °C. The mean temperature of the incubator used for the negative controls was 30.0 (± 1) °C.
Temperature profile of a typical experiment running in total over 8 h. The temperatures (centrifuge, biological incubator, and room temperature) were monitored using iButton® temperature data loggers in all experiments. Centrifuge was pre-run at 800×g to reach the required temperature (30 °C) quickly. Then, the experiments were performed at 200×g
Influence of temperature on cell viability
A multi-parameter kit was used for testing apoptosis progression at different time points under two different temperatures (30 °C vs. 37 °C). Both, the experimental group at 30 °C and the negative control group at 37 °C provided identical findings (Fig. 3). There were no signs of apoptosis detectable up to 4 h of incubation at both, 30 °C and 37 °C. Hoechst staining showed that all cells presented with round shape and intact nuclei. TMRE staining revealed that cells had undisrupted mitochondrial membranes. Testing with Annexin V was negative in all samples irrespective of cultivation temperature. In contrast, the apoptosis-positive controls showed distinctly different staining patterns after 24 h of apoptosis induction with 4 mM dithiothreitol (Fig. 3). In both human PDL-derived fibroblasts (HPDFs) and alveolar bone-derived osteoblasts (HABOs), cells showed clearly signs of apoptosis: condensed and fragmented nuclei, abolished or diminished membrane potential, and positive Annexin V staining.
Apoptosis induction at 30 °C in comparison to negative control (37 °C, 5% CO2) and apoptosis-positive controls (4 mM dithiothreitol for 24 h) in HPDF and HABO. Cells were stained with TMRE/Hoechst Dye and Annexin V-FITC. From left to right: phase contrast, Annexin V, mitochondrial membrane potential, DNA fragmentation and their overlay images are shown. Scale bar: 200 μm
Gene expression
Gene expression results following mono- and co-culture of HPDFs are shown in Fig. 4, and the results derived from HABOs are presented in Fig. 5.
Gene expression of TNFA, PTGS2, and IL6 in mono-cultured (a, c, e) and co-cultured (b, d, f) HPDFs after 1, 2, and 4 h of CF at 47.4 g/cm2. The ΔΔCq method was applied and B2M was used as the reference gene. Expression level of control was defined as 1. Values were presented as mean ± SEM (*pcorr < 0.017, **pcorr < 0.003, ***pcorr < 0.0003). Black bars indicate control; white bars indicate compressive force
Gene expression of TNFA, PTGS2, and IL6 in mono-cultured (a, c, e) and co-cultured (b, d, f) HABOs after 1, 2, and 4 h of CF at 47.4 g/cm2. The ΔΔCq method was applied and B2M was used as the reference gene. Expression level of control was defined as 1. Values were presented as mean ± SEM (*pcorr < 0.017, **pcorr < 0.003, ***pcorr < 0.0003). Black bars indicate control; white bars indicate compressive force
HPDFs
A slight upregulation of TNFA gene expression in mono-culture was observed after 1, 2, and 4 h of CF application (Fig. 4a). In comparison to unloaded controls, the relative PTGS2 gene expression showed a significant increase after 1 h (2.25 ± 0.25, p = 0.001) and 2 h (4.86 ± 0.47, p < 0.001) but not at 4 h of CF (Fig. 4c). A significant upregulation of the IL6 gene expression was found in samples that have been exposed to CF after 1 h (p < 0.001) and 2 h (p < 0.001) but not after 4 h (Fig. 4e).
In co-culture, no significant changes in TNFA gene expression were found between cells exposed to CF and negative control samples (Fig. 4b). However, relative gene expression of PTGS2 was increased after 1 h (p = 0.005) of CF and significantly reduced after 4 h of CF (p < 0.001) in comparison to that of the control samples (Fig. 4d). IL6 gene expression showed no significant differences between cells with and without exposure to CF after 1 h and 2 h, but was significantly decreased at 4 h (p = 0.001) (Fig. 4f).
HABOs
In mono-culture, CF induced a stronger TNFA gene expression (4.0 ± 0.38, p < 0.001) in comparison to control samples after 1 h only (Fig. 5a). PTGS2 gene expression was significantly stronger under CF in these cells and showed a constant increase with time (Fig. 5c). Similar was observed for IL6 gene expression indicating only a small but significant increase in comparison to unloaded controls after 1 h (p < 0.001) and 2 h (p < 0.001) of CF which was even stronger after 4 h (5.15 ± 0.46, p < 0.001) (Fig. 5e).
In co-culture, relative gene expression of TNFA showed a slight increase after 2 h (p = 0.002) of CF but remained unchanged in comparison to negative control samples at 1 h and 4 h (Fig. 5b). CF induced a stronger PTGS2 gene expression after 1 h (p < 0.001) and 2 h (p < 0.001) but not at 4 h (Fig. 5d). Considering IL6 gene expression CF caused a significant increase at 2 h (2.91 ± 0.46, p = 0.001) and 4 h (1.39 ± 0.11, p = 0.005) (Fig. 5f).
Effects of CF on TNF, PGE2, and IL6 protein expression
Protein expression of PGE2 and IL6 in mono- and co-cultured HPDFs and HABOs subjected to CF is shown in Fig. 6. Protein levels of TNF were below the detection limit of the ELISA kit in all samples and are therefore not shown here. To discriminate between both co-culture configurations (HPDFs in the well and HABOs in the insert, or vice versa), the cell type in the well is named.
PGE2 and IL6 concentrations of mono-cultured (a–d) and co-cultured (e–h) HPDFs (a, b, e, f) and HABOs (c, d, g, h) after 1, 2, and 4 h of CF at 47.4 g/cm2 were measured using ELISA. The schematic drawings depict the cell type seeded in the well in both mono- and co-culture setups (see also Fig. 1). Control groups were not subjected to CF. Values are presented as mean ± SEM (*pcorr < 0.017, **pcorr < 0.003, ***pcorr < 0.0003). Black bars indicate control; white bars indicate compressive force
Application of CF in mono-cultured cells
In HPDFs, PGE2 was found at higher concentrations in cells exposed to CF than in controls at any time point, but the difference reached significance at 1 h (p = 0.016) and 4 h (p = 0.004) only (Fig. 6a). In HABOs, PGE2 was increased after 1 h and 2 h of CF application (p = 0.038), being not significant after correction for multiple testing in both cases. A non-significant decrease after 4 h of CF was observed (Fig. 6c). The IL6 protein levels in HPDFs were significantly higher than in controls only after 2 h (p = 0.002) of CF application (Fig. 6b). No significant difference was found in HABOs at any time point (Fig. 6d).
Application of CF in co-cultured cells
In HPDFs, the concentration of PGE2 was significantly smaller in samples treated with CF than in controls after 1 h (p = 0.002) and 4 h (p = 0.002) (Fig. 6e). In HABOs, the PGE2 concentration significantly increased after 2 h (p = 0.019; not significant after Bonferroni’s correction) of CF, but was smaller after 1 h and 4 h of CF (Fig. 6g). Concerning IL6, a smaller amount was found in HPDFs treated with CF at 1 h (p = 0.026; not significant after Bonferroni’s correction) (Fig. 6f). Also, in HABOs, the IL6 protein concentration was smaller in samples after CF application at 1 h (p = 0.041) of CF, but significantly higher at 2 h (p = 0.015) (Fig. 6h).
Comparison of the force-induced changes in gene and protein expression according to culture conditions
The changes in gene expression induced by CF showed considerable dependency on the culture conditions. With exception of the TNFA gene expression at 1 h co-culture of both tested cell types (Table 2) attenuated the CF-induced augmentation of TNFA, PTGS2, and IL6 gene expression as well as the PGE2 and IL6 protein expression in HPDFs in comparison to mono-culture (Table 2). However, these differences between mono- and co-culture reached significance not in all cases. A comparable reduction of the force-induced changes in gene and protein expression was also found in HABOs, but not for those samples that have been exposed to CF for 2 h and for the protein expression of IL6 at 4 h (Table 2).
Discussion
The effect of mechanical force on bone remodeling during orthodontic tooth movement is yet mostly studied using mono-cultured cells in vitro [3, 6]. However, in vivo, the periodontal tissue represents a heterogeneous population of different cell types which might be functionally closely interrelated [17]. During orthodontic tooth movement, these cells are subjected to mechanical force simultaneously leading to more or less intensive intercellular communication in this complex tissue microenvironment [19, 27]. Therefore, co-culture of various cell types together seems more appropriate since it reflects the real physiological conditions much closer and, in addition, enables the investigation of intercellular communication in vitro [20]. In this study, indirect-contact co-culture was applied to show if there exists a kind of communication between various periodontal cell types, i.e., human PDL-derived fibroblasts (HPDFs) and alveolar bone-derived osteoblasts (HABOs) when subjected to static compressive force (CF) application. The indirect-contact co-culture model as used herein separated the HPDFs from the HABOs with a porous membrane but kept both cell types under common growth conditions allowing signaling molecules and other mediators, e.g., cytokines, growth factors, and metabolites to pass the membrane unrestrictedly.
To carry out orthodontic tooth movement, the effects of CF inducing controlled bone resorption in the intended moving direction is essential [28]. Several methods have been described to apply mechanical forces in vitro on cell cultures, e.g., hydrostatic pressure [29], direct weight application [15], microgravity [30], or centrifugation [23, 31,32,33,34]. Only centrifugation applies equal amounts of CF to different types of cells growing in indirect co-culture and allows individual gene expression analysis of each cell type afterwards.
Since biological systems are temperature-sensitive, human cell cultivation is commonly done at a temperature of 37 °C. Cell culture at temperatures below 37 °C over longer periods of time will most probably induce apoptosis and cell death. Although the use of centrifugation has been widely accepted as method for CF application, most of the previous studies were done at room temperature (i.e., 20–24 °C) [23, 31,32,33]. Herein, we were able to create a stable temperature of 30 °C in the experimental setup. The influence of this temperature on cell viability and apoptosis induction was compared to cells that were incubated at 37 °C. Our results showed no apoptosis induction done after cell cultivation at 30 °C for up to 4 h in comparison to incubation at 37 °C. This observation is in line with previous studies on periodontal ligament fibroblast viability after traumatic avulsion of permanent teeth [35]. Hence, the cultivation of human cells at 30 °C seems to be feasible during in vitro experiments for up to 4 h.
The inflammatory reaction of periodontal tissue cells during application of orthodontic forces is mediated by various signaling molecules [5]. In this study, we found that the gene expression of the pro-inflammatory and tissue remodeling mediators, TNFA, PTGS2, and IL6, were considerably influenced by CF application, depending on the duration of exposure. Roughly, the gene expression of TNFA, PTGS2, and IL6 as well as the protein expression of PGE2 and IL6 was stimulated in mono-cultured HPDFs and HABOs by the presence of CF. Similar findings have been reported previously showing that CF induces a strong amplification of pro-inflammatory and bone resorbing mediators in various periodontal cell types [3, 7, 15, 18].
Intriguingly, when HPDFs and HABOs are co-cultured together, the mechanically induced upregulation of gene expression shows considerable changes as compared to the mono-cultured cells. When interpreting these changes, one has to consider also the strong overall increase in the PGE2 and IL6 expression as found in the ELISA experiments for HPDF cells that have been cultured alone or together with HABOs. So far, the specific reason remains unclear, but this increase might probably reflect the influence of different culture media as used in mono- and co-culture experiments with HPDFs due to biological reasons.
Several recent studies [19, 36, 37] have demonstrated that co-culture of different cell types has the potential to induce changes in the cellular phenotype including the expression of various mediators. For example, co-culture with an osteoblastic cell line led to a strong increase of the expression of the pro-inflammatory cytokines interleukin-6 and interleukin-8 in oral keratinocytes [36]. Moreover, co-culture of both cell types resulted in a stronger activation of osteoclasts than each cell type alone. Most likely, these observations are mediated by the presence of elevated osteopontin levels. Another study demonstrated that co-cultured osteoblasts are able to induce the differentiation of adipose-derived stem cells into osteoblasts [37]. Using co-cultures of dental pulp cells and periodontal ligament cells, it was shown that the presence of pulp cells enhances the expression of RANKL, PGE2, and substance P in periodontal ligament cells [19].
The mechanical force induced a significantly increased transcription of the IL6 gene in osteoblasts after 4 h but, on the contrary, led to an increased protein level of IL6. Yet one can only speculate on the specific reasons. As shown in rat alveolar type II cells, cyclic mechanical forces differentially affect the intracellular levels and the secretion of TNF and interleukin 6 which is further modulated according to the CO2 concentration [38]. Hence, the collaborative influence of compressive force and the cross talk between the two cell types might reduce the transcription of the IL6 gene and increase the enhanced secretion of intracellular IL6 in osteoblasts.
The changes of the force-induced upregulation of gene expression were dependent upon the specific placement of cells within the experimental setup, i.e., the insert or the bottom of the well. When placing the HPDFs in the bottom of the well, the force-induced enhancement of the expression of the tested marker molecules is strongly attenuated in presence of HABOs. On the contrary, when HBAOs are placed at the bottom of the well, the force-induced increasing expression of some of the marker molecules received further enhancement at several time points. Although this study did not specifically refer to this issue, one might speculate that each cell type exerts different effects on the second cell type. During CF application, the force vector \( \overrightarrow{F} \) is perpendicular to the growth plane of both cells [25, 34]. Hence, functional effective molecules released by the cells seeded into the insert might migrate along the force vector during centrifugation to the second cell type at the bottom of the well leading to different phenotypic changes. This might at least partially reflect the real clinical situation during orthodontic tooth movements. At the side of the tooth root in line to the movement, the pressure as induced by the orthodontic appliance leads to controlled resorption of osseous tissue. According to the present results, the neighboring cells of the periodontal tissue probably might enhance the osseous tissue resorptive activity by inducing IL 6 and TNF in osteoblasts. Conversely, the desmodontal connective tissue remains stable during the entire orthodontic tooth movement and, thus, remains unaffected [39]. Hence, the osteoblasts adjacent to the periodontal ligament cells mediate the attenuation of tissue resorbing signals.
It must be noted, that intercellular communication is a complex bidirectional, spatial, and temporal process. Taken together, several studies and the present findings show considerable differences between mono- and co-cultured cells in inflammatory and bone remodeling mediators’ expression. This indicates a kind of paracrine signaling pathway [40,41,42] between HPDFs and HABOs. Herein, this communication results in a strong downregulation of the TNFA, PTGS2, and IL6 gene expressions during CF application. To identify signaling molecules involved in regulation of mechanical force transmission, further studies are necessary. In this context, the TNF signaling pathway (KEGG [43] pathway no. ko04668) might be an attractive candidate for further studies [3].
Conclusions
Our data show that CF enhances the gene expression of TNFA, PTGS2, and IL6 and, in most instances, also, the protein activity of PGE2 and IL6 in mono-cultured HPDFs and HABOs. Co-culture of HPDFs and HABOs seems to result in a repressive effect on the force-induced increase of both: gene and protein expressions. This indicates a kind of intercellular communication between both cell types in vitro.
References
Nanci A, Bosshardt DD (2006) Structure of periodontal tissues in health and disease. Periodontol 2000(40):11–28. https://doi.org/10.1111/j.1600-0757.2005.00141.x
Marchesan JT, Scanlon CS, Soehren S, Matsuo M, Kapila YL (2011) Implications of cultured periodontal ligament cells for the clinical and experimental setting: a review. Arch Oral Biol 56(10):933–943. https://doi.org/10.1016/j.archoralbio.2011.03.003
Koyama Y, Mitsui N, Suzuki N, Yanagisawa M, Sanuki R, Isokawa K, Shimizu N, Maeno M (2008) Effect of compressive force on the expression of inflammatory cytokines and their receptors in osteoblastic Saos-2 cells. Arch Oral Biol 53(5):488–496. https://doi.org/10.1016/j.archoralbio.2007.12.004
Nakao K, Goto T, Gunjigake KK, Konoo T, Kobayashi S, Yamaguchi K (2007) Intermittent force induces high RANKL expression in human periodontal ligament cells. J Dent Res 86(7):623–628. https://doi.org/10.1177/154405910708600708
Li Y, Li M, Tan L, Huang S, Zhao L, Tang T, Liu J, Zhao Z (2013) Analysis of time-course gene expression profiles of a periodontal ligament tissue model under compression. Arch Oral Biol 58(5):511–522. https://doi.org/10.1016/j.archoralbio.2012.10.006
Jacobs C, Walter C, Ziebart T, Grimm S, Meila D, Krieger E, Wehrbein H (2014) Induction of IL-6 and MMP-8 in human periodontal fibroblasts by static tensile strain. Clin Oral Investig 18(3):901–908. https://doi.org/10.1007/s00784-013-1032-1
Mayahara K, Kobayashi Y, Takimoto K, Suzuki N, Mitsui N, Shimizu N (2007) Aging stimulates cyclooxygenase-2 expression and prostaglandin E2 production in human periodontal ligament cells after the application of compressive force. J Periodontal Res 42(1):8–14. https://doi.org/10.1111/j.1600-0765.2006.00885.x
Kim SJ, Park KH, Park YG, Lee SW, Kang YG (2013) Compressive stress induced the up-regulation of M-CSF, RANKL, TNF-α expression and the down-regulation of OPG expression in PDL cells via the integrin-FAK pathway. Arch Oral Biol 58(6):707–716. https://doi.org/10.1016/j.archoralbio.2012.11.003
Osta B, Benedetti G, Miossec P (2014) Classical and paradoxical effects of TNF-α on bone homeostasis. Front Immunol 5(48). https://doi.org/10.3389/fimmu.2014.00048
Bertolini DR, Nedwin GE, Bringman TS, Smith DD, Mundy GR (1986) Stimulation of bone resorption and inhibition of bone formation in vitro by human tumour necrosis factors. Nature 319(6053):516–518. https://doi.org/10.1038/319516a0
De Benedetti F, Rucci N, Del Fattore A, Peruzzi B, Paro R, Longo M, Vivarelli M, Muratori F, Berni S, Ballanti P, Ferrari S, Teti A (2006) Impaired skeletal development in interleukin-6-transgenic mice: a model for the impact of chronic inflammation on the growing skeletal system. Arthritis Rheum (Munch) 54(11):3551–3563. https://doi.org/10.1002/art.22175
Baker PJ, Dixon M, Evans RT, Dufour L, Johnson E, Roopenian DC (1999) CD4+ T cells and the proinflammatory cytokines gamma interferon and interleukin-6 contribute to alveolar bone loss in mice. Infect Immun 67(6):2804–2809
Yang X, Ricciardi BF, Hernandez-Soria A, Shi Y, Pleshko Camacho N, Bostrom MP (2007) Callus mineralization and maturation are delayed during fracture healing in interleukin-6 knockout mice. Bone 41(6):928–936. https://doi.org/10.1016/j.bone.2007.07.022
Balto K, Sasaki H, Stashenko P (2001) Interleukin-6 deficiency increases inflammatory bone destruction. Infect Immun 69(2):744–750. https://doi.org/10.1128/IAI.69.2.744-750.2001
Kanzaki H, Chiba M, Shimizu Y, Mitani H (2002) Periodontal ligament cells under mechanical stress induce osteoclastogenesis by receptor activator of nuclear factor in κB ligand up-regulation via prostaglandin E2 synthesis. J Bone Miner Res 17(2):210–220. https://doi.org/10.1359/jbmr.2002.17.2.210
Shimizu N, Ozawa Y, Yamaguchi M, Goseki T, Ohzeki K, Abiko Y (1998) Induction of COX-2 expression by mechanical tension force in human periodontal ligament cells. J Periodontol 69(6):670–677. https://doi.org/10.1902/jop.1998.69.6.670
Li J, Wan Z, Liu H, Li H, Liu L, Li R, Guo Y, Chen W, Zhang X, Zhang X (2013) Osteoblasts subjected to mechanical strain inhibit osteoclastic differentiation and bone resorption in a co-culture system. Ann Biomed Eng 41(10):2056–2066. https://doi.org/10.1007/s10439-013-0810-x
Römer P, Köstler J, Koretsi V, Proff P (2013) Endotoxins potentiate COX-2 and RANKL expression in compressed PDL cells. Clin Oral Investig 17(9):2041–2048. https://doi.org/10.1007/s00784-013-0928-0
Morikawa T, Matsuzaka K, Nakajima K, Yasumura T, Sueishi K, Inoue T (2016) Dental pulp cells promote the expression of receptor activator of nuclear factor-κB ligand, prostaglandin E2 and substance P in mechanically stressed periodontal ligament cells. Arch Oral Biol 70:158–164. https://doi.org/10.1016/j.archoralbio.2016.06.021
Hatherell K, Couraud PO, Romero IA, Weksler B, Pilkington GJ (2011) Development of a three-dimensional, all-human in vitro model of the blood-brain barrier using mono-, co-, and tri-cultivation Transwell models. J Neurosci Methods 199(2):223–229. https://doi.org/10.1016/j.jneumeth.2011.05.012
Somerman MJ, Archer SY, Imm GR, Foster RA (1988) A comparative study of human periodontal ligament cells and gingival fibroblasts in vitro. J Dent Res 67(1):66–70. https://doi.org/10.1177/00220345880670011301
Ng KW, Schantz J-T (2010) A manual for primary human cell culture. Manuals in Biomedical Research, vol 6, 2nd edn. World Scientific, New Jersey
Redlich M, Palmon A, Zaks B, Geremi E, Rayzman S, Shoshan S (1998) The effect of centrifugal force on the transcription levels of collagen type I and collagenase in cultured canine gingival fibroblasts. Arch Oral Biol 43(4):313–316. https://doi.org/10.1016/S0003-9969(97)00108-8
Redlich M, Roos H, Reichenberg E, Zaks B, Grosskop A, Bar Kana I, Pitaru S, Palmon A (2004) The effect of centrifugal force on mRNA levels of collagenase, collagen type-I, tissue inhibitors of metalloproteinases and β-actin in cultured human periodontal ligament fibroblasts. J Periodontal Res 39(1):27–32. https://doi.org/10.1111/j.1600-0765.2004.00700.x
Redlich M, Asher Roos H, Reichenberg E, Zaks B, Mussig D, Baumert U, Golan I, Palmon A (2004) Expression of tropoelastin in human periodontal ligament fibroblasts after simulation of orthodontic force. Arch Oral Biol 49(2):119–124. https://doi.org/10.1016/j.archoralbio.2003.08.002
Xiang XY, Yang XC, Su J, Kang JS, Wu Y, Xue YN, Dong YT, Sun LK (2016) Inhibition of autophagic flux by ROS promotes apoptosis during DTT-induced ER/oxidative stress in HeLa cells. Oncol Rep 35(6):3471–3479. https://doi.org/10.3892/or.2016.4725
Krishnan V, Davidovitch Z (2006) Cellular, molecular, and tissue-level reactions to orthodontic force. Am J Orthod Dentofac Orthop 129(4):469.e1–469.32. https://doi.org/10.1016/j.ajodo.2005.10.007
Wise GE, King GJ (2008) Mechanisms of tooth eruption and orthodontic tooth movement. J Dent Res 87(5):414–434. https://doi.org/10.1177/154405910808700509
Nakago-Matsuo C, Matsuo T, Nakago T (1996) Intracellular calcium response to hydraulic pressure in human periodontal ligament fibroblasts. Am J Orthod Dentofac Orthop 109(3):244–248. https://doi.org/10.1016/S0889-5406(96)70147-6
Carmeliet G, Nys G, Stockmans I, Bouillon R (1998) Gene expression related to the differentiation of osteoblastic cells is altered by microgravity. Bone 22(5 Suppl):139S–143S. https://doi.org/10.1016/S8756-3282(98)00007-6
Inoue H, Nakamura O, Duan Y, Hiraki Y, Sakuda M (1993) Effect of centrifugal force on growth of mouse osteoblastic MC3T3-E1 cells in vitro. J Dent Res 72(9):1351–1355. https://doi.org/10.1177/00220345930720091601
Fitzgerald J, Hughes-Fulford M (1999) Mechanically induced c-fos expression is mediated by cAMP in MC3T3-E1 osteoblasts. FASEB J 13(3):553–557
Theilig C, Bernd A, Leyhausen G, Kaufmann R, Geurtsen W (2001) Effects of mechanical force on primary human fibroblasts derived from the gingiva and the periodontal ligament. J Dent Res 80(8):1777–1780. https://doi.org/10.1177/00220345010800081901
Baumert U, Golan I, Becker B, Hrala BP, Redlich M, Roos HA, Palmon A, Reichenberg E, Müßig D (2004) Pressure simulation of orthodontic force in osteoblasts: a pilot study. Orthod Craniofac Res 7(1):3–9. https://doi.org/10.1046/j.1601-6335.2003.00270.x
Souza BD, Lückemeyer DD, Felippe WT, Simões CM, Felippe MC (2010) Effect of temperature and storage media on human periodontal ligament fibroblast viability. Dent Traumatol 26(3):271–275. https://doi.org/10.1111/j.1600-9657.2010.00886.x
Teixeira LN, de Castro Raucci LM, Alonso GC, Coletta RD, Rosa AL, de Oliveira PT (2016) Osteopontin expression in co-cultures of human squamous cell carcinoma-derived cells and osteoblastic cells and its effects on the neoplastic cell phenotype and osteoclastic activation. Tumour Biol 37(9):12371–12385. https://doi.org/10.1007/s13277-016-5104-0
Liu DC, Yang XN, Huang CZ, Sun YG, Dai XM (2016) Experimental study on co-culturing adipose-derived stem cells with osteoblasts under different conditions. Eur Rev Med Pharmacol Sci 20(17):3535–3543
Dixon DL, Barr HA, Bersten AD, Doyle IR (2008) Intracellular storage of surfactant and proinflammatory cytokines in co-cultured alveolar epithelium and macrophages in response to increasing CO2 and cyclic cell stretch. Exp Lung Res 34(1):37–47. https://doi.org/10.1080/01902140701807928
Lindskog-Stokland B, Wennström JL, Nyman S, Thilander B (1993) Orthodontic tooth movement into edentulous areas with reduced bone height. An experimental study in the dog. Eur J Orthod 15(2):89–96. https://doi.org/10.1093/ejo/15.2.89
Kim CH, You L, Yellowley CE, Jacobs CR (2006) Oscillatory fluid flow-induced shear stress decreases osteoclastogenesis through RANKL and OPG signaling. Bone 39(5):1043–1047. https://doi.org/10.1016/j.bone.2006.05.017
Kook SH, Son YO, Hwang JM, Kim EM, Lee CB, Jeon YM, Kim JG, Lee JC (2009) Mechanical force inhibits osteoclastogenic potential of human periodontal ligament fibroblasts through OPG production and ERK-mediated signaling. J Cell Biochem 106(6):1010–1019. https://doi.org/10.1002/jcb.22086
Mayahara K, Yamaguchi A, Takenouchi H, Kariya T, Taguchi H, Shimizu N (2012) Osteoblasts stimulate osteoclastogenesis via RANKL expression more strongly than periodontal ligament cells do in response to PGE2. Arch Oral Biol 57(10):1377–1384. https://doi.org/10.1016/j.archoralbio.2012.07.009
Manyam G, Birerdinc A, Baranova A (2015) KPP: KEGG pathway painter. BMC Syst Biol 9(Suppl 2):S3. https://doi.org/10.1186/1752-0509-9-S2-S3
Acknowledgments
We thank Lisa Reuther and Christine Schreindorfer (both: Department of Orthodontics and Dentofacial Orthopedics, LMU Munich) for their assistance in the laboratory and Dr. Julia Diegelmann and Brigitte Hackl (both: Department of Conservative Dentistry and Periodontology, LMU Munich) for their technical support.
Funding
J. S. was supported by a grant from the China Scholarship Council (CSC, 201608080067).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. This article does not contain any studies with animals performed by any of the authors.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Shi, J., Baumert, U., Folwaczny, M. et al. Influence of static forces on the expression of selected parameters of inflammation in periodontal ligament cells and alveolar bone cells in a co-culture in vitro model. Clin Oral Invest 23, 2617–2628 (2019). https://doi.org/10.1007/s00784-018-2697-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00784-018-2697-2