Abstract
Chlorite dismutase is a unique heme enzyme that catalyzes the conversion of chlorite to chloride and molecular oxygen. The enzyme is highly specific for chlorite but has been known to bind several anionic and neutral ligands to the heme iron. In a pH study, the enzyme changed color from red to green in acetate buffer pH 5.0. The cause of this color change was uncovered using UV–visible and EPR spectroscopy. Chlorite dismutase in the presence of acetate showed a change of the UV–visible spectrum: a redshift and hyperchromicity of the Soret band from 391 to 404 nm and a blueshift of the charge transfer band CT1 from 647 to 626 nm. Equilibrium binding titrations with acetate resulted in a dissociation constant of circa 20 mM at pH 5.0 and 5.8. EPR spectroscopy showed that the acetate bound form of the enzyme remained high spin S = 5/2, however with an apparent change of the rhombicity and line broadening of the spectrum. Mutagenesis of the proximal arginine Arg183 to alanine resulted in the loss of the ability to bind acetate. Acetate was discovered as a novel ligand to chlorite dismutase, with evidence of direct binding to the heme iron. The green color is caused by a blueshift of the CT1 band that is characteristic of the high spin ferric state of the enzyme. Any weak field ligand that binds directly to the heme center may show the red to green color change, as was indeed the case for fluoride.
Graphic abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chlorite dismutase is an essential enzyme in (per)chlorate-respiring bacteria [1, 2]. The enzyme catalyzes the disproportionation of the toxic compound chlorite ClO2−, the endproduct of (per)chlorate respiration, into harmless Cl− and O2. This heme b containing enzyme is unique as it catalyzes O=O bond formation which is otherwise only catalyzed by the oxygen-evolving manganese cluster of photosystem II [3]. Chlorite dismutase has been extensively studied structurally and spectroscopically [4,5,6,7]. However, to date, the precise mechanism of the enzyme remains elusive.
Genomic studies have revealed two distinct classes of chlorite dismutase (Cld), penta- and hexameric Cld or dimeric Cld [8]. Furthermore, phylogenetic and structural relationships exist with the coproheme decarboxylase HemQ and the dye decolorizing peroxidases DyP [9, 10]. HemQ is involved in heme biosynthesis in bacteria that belong to the Firmicutes and Actinobacteria. DyPs are enzymes with a broad substrate scope and are unrelated to other types of peroxidases. Together all these protein clades form the peroxidase-chlorite dismutase superfamily [11].
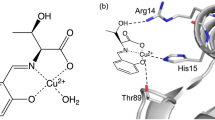
The dimeric Cld enzymes are structurally (subunit structure and interface, conformational as well as thermal stability) and spectroscopically different. Dimeric Clds are from bacteria that are not chlorate-respiring but can catalyze chlorite conversion. The biological function of these enzymes is not clear, although the organisms contain nitrate reductases that may reduce chlorate non-productively to chlorite [12, 13]. Trp156 close to the heme is highly conserved in pentameric Cld, but not in dimeric Cld. The crystal structure of recombinantly expressed chlorite dismutase from Azospira oryzae (AoCld) shows that it is a homohexamer (although likely a pentamer in solution), in which each monomer consists of two domains with a ferredoxin-like fold. The active site comprises a heme b cofactor with an axial His170 and a proximal Arg183 (Fig. 1) [6]. The conserved Arg183 adopts a so-called “in” position, where the guanidinium group is pointing towards the heme group, or “out” position, where the guanidinium group is pointing towards the entry of the substrate channel.
Crystal structure of AoCld (PDB 2vxh). a The crystal structure discloses the hexameric form of AoCld [6]. The heme cofactor and important residues are depicted as sticks. b The monomeric form of AoCld is showing the secondary structure elements. c The active site architecture of AoCld with bound thiocyanate at the active site. Figures were generated using PyMOL [49]
The UV–visible spectrum of AoCld is characteristic of a penta-coordinated high spin (HS) ferric heme protein. The Soret peak is broad and has a peak maximum at 391 nm. The Q bands are broad featured around 510 nm and the charge transfer CT1 band is found at 647 nm. The sixth coordination position is available to the substrate and different ligands. Cld has been found to bind (predominantly anionic) ligands: fluoride, cyanide, azide, thiocyanate, nitrite and imidazole [4, 6, 14, 15]. The R183A mutation has been found to abolish azide and fluoride binding, but not imidazole in DaCld, which suggests that Arg183 stabilizes anionic ligand binding by hydrogen bond donation [16].
The activity and spectroscopic properties of Cld are highly pH-dependent [4, 16, 17]. Upon examination of the pH dependence of the activity and stability of AoCld we discovered that the enzyme solution became green-colored in acetate buffer pH 5.0 (Fig. S1). The color change was reversible after buffer exchange due to the reversible binding of acetate as a ligand to the heme iron in AoCld. To our knowledge, the binding of acetate directly to the heme iron is almost unique to Cld. The UV–visible and EPR spectroscopic properties of the green-colored acetate bound AoCld are reported and compared to the fluoride and imidazole adducts of AoCld.
Materials and methods
Chemicals
Antibiotics (kanamycin and chloramphenicol), hemin and IPTG procured from Sigma. Buffer components and purification reagents were purchased from Sigma and Merck.
Protein production and purification
The strains and plasmids that were used are given in Table S1. AoCld was recombinantly expressed in E. coli using the pET28a-AoCld construct in E. coli BL21(DE3) pLysS as described elsewhere [18]. The expression strain was cultivated in TB medium containing 50 µg mL−1 kanamycin, 25 μg mL−1 chloramphenicol and 60 μg mL−1 hemin (pre-dissolved in 1.4 N NaOH) in a 15 L fermenter (Applikon) at 37 °C. The fermenter was stirred at 750 rpm with an air inflow of 5 L min−1. Expression was induced by the addition of 119 μg mL−1 IPTG when the culture reached an OD600 = 0.5. The cultivation temperature was reduced from 37 to 25 °C after induction. The cells were harvested by centrifugation 7 h after induction using a Sorvall centrifuge at 17,000g for 10 min at 4 °C. The cell pellet was washed twice using 20 mM Tris–HCl pH 7.5 containing 500 mM NaCl and 50 mM imidazole (Buffer A). The resulting AoCld containing cells were resuspended in Buffer A containing additionally 1 mM PMSF, 1 mM MgSO4, 200 mg L−1 lysozyme and 10 μg mL−1 DNase. The cell suspension was homogenized with a Homogenizer RW16 (IKA) and subsequently disrupted using a cell disruptor (Constant Systems) operating at 1.5 kbar. The cell-free extract (CFE) was obtained as the supernatant after centrifugation using a Sorvall centrifuge at 17,000g for 90 min at 4 °C. The CFE was filter sterilized using a 0.22 μm Steritop filter (Millipore) before protein purification.
AoCld was purified from the CFE using IMAC chromatography as the recombinant protein contained an N-terminal His6 tag. A 150 mL Ni Sepharose 6 Fast Flow column (GE Healthcare) was equilibrated with 4 column volumes (CV) buffer A using an NGC medium pressure liquid chromatography system (Bio-Rad) at a flow rate of 3 mL min−1. 1.5 L CFE was loaded onto the column using a flow rate of 0.7 mL min−1 overnight, and subsequently, the column was washed using 1.5 CV buffer A at a flow rate of 3 mL min−1. AoCld was eluted from the column using a linear gradient from 0–100% 20 mM Tris–HCl pH 7.5 containing 150 mM NaCl and 500 mM imidazole (buffer B) in 4 CV and 2 CV 100% buffer B. AoCld started to elute at circa 325 mM imidazole.
AoCld was subsequently desalted to remove excess imidazole with HiTrap Desalting columns (GE Healthcare) using the NGC system (Bio-Rad) with 20 mM Tris–HCl pH 7.5 (flow rate 4 mL min−1). The buffer was exchanged to 50 mM KPi pH 7.0 using a gravitation flow desalting column PD10 (GE Healthcare) according to the manufacturer’s instructions. To further remove bound imidazole the protein was extensively dialyzed against 100 mM KPi pH 7.0 to a dilution factor of 650 with a dialysis cycle of a minimum 8 h using a dialysis membrane with a 3.5 kDa cutoff and a diameter of 16 mm (Visking dialysis tubing, Medicell Membranes) at 5 °C. The purified AoCld was concentrated to 8 mg mL−1 using a centrifugal ultrafiltration device with a 30 kDa cutoff (Amicon filter, Millipore) and stored at − 80 °C until further use. The concentration of pure AoCld was determined spectrophotometrically using ε280 = 52.41 mM−1 cm−1 (monomer concentration).
AoCld R183A was produced with the QuikChange kit (Agilent) using the mutagenic primers AoCld-R183A-F 5′-CTGGTGAATGTCAAAGCGAAGTTGTATCAC-3′ and AoCld-R183A-R 5′-GTGATACAACTTCGCTTTGACATTCACCAG -3′ with the mutagenic codon underlined. E. coli pET28a AoCld-R183A was cultivated in 1 L TB medium as described above. The enzyme was purified using the same procedure as described above for the WT enzyme.
Biochemical characterization
The protein concentration was determined according to the Bicinchoninic acid method using the BCA protein assay kit (Uptima, Interchim) according to the manufacturer’s instructions [19]. The protein purity was examined using SDS-PAGE using the Criterion XT gel system (Bio-Rad). UV–visible spectra of AoCld were measured using the Cary 60 spectrophotometer (Agilent) at 21 °C. AoCld activity was measured polarographically as oxygen is produced during the reaction using the Clark-type oxygen electrode (type 5331, YSI Life Science) in 100 mM KPi pH 7.0 using 1 mM sodium chlorite as substrate at 20 °C.
Equilibrium binding titrations
Equilibrium binding titrations were performed using acetate, fluoride, and imidazole as ligands by monitoring the change of the Soret band. The UV–visible spectrum of AoCld was monitored and 0–180 mM sodium acetate was titrated to 10 µM AoCld in 100 mM citrate–phosphate buffer pH 5.0 and 100 mM KPi pH 5.8. For acetate as a ligand, the absorbance difference between the Soret maximum at 404 nm and the isosbestic point at 396 nm was plotted against the unbound ligand concentration. For the titration with fluoride as a ligand 0–120 mM NaF was titrated against 10 μM AoCld in 100 mM KPi pH 7.0 and the absorbance difference between the Soret maximum at 400 nm and the isosbestic point at 390 nm was plotted. For imidazole 0–1 mM ligand was titrated against 10 µM AoCld in 100 mM KPi pH 5.0, 100 mM KPi pH 5.8, 100 mM KPi pH 7.0 and 100 mM Tris buffer pH 8.0, and the absorbance difference between 412 nm and the isosbestic point at 404 nm was plotted. The data were fitted to Eq. 1.
KD is the dissociation constant. [L] represents the unbound ligand concentration, which was assumed equal to the total ligand concentration for acetate and fluoride. In the case of imidazole, the unbound ligand concentration was corrected for the bound ligand concentration using the experimentally determined extent of saturation and the total enzyme concentration.
EPR spectroscopy
EPR measurements were performed using a Bruker EMXplus 9.5 spectrometer and the following conditions: 9.402 GHz microwave frequency, 0.2 mW microwave power, 100 kHz modulation frequency, 10 Gauss modulation amplitude and a temperature of 20 K, unless stated otherwise. The low temperature was maintained by boiling liquid helium and cold helium vapor was passed through a double-wall quartz glass tube, which was mounted and fitted in the rectangular cavity [20, 21]. Samples were prepared containing 75 μM (monomer concentration) AoCld in 100 mM KPi pH 7.0 containing ligands or 100 mM acetate buffer pH 5.0 to afford a final volume of 200 µL.
Results
Bound imidazole was difficult to remove during AoCld purification
Overexpressed AoCld was purified through a Nickel Sepharose column with elution buffer of 20 mM Tris–HCl and 150 mM NaCl, 500 mM imidazole pH 7.5. The high imidazole concentration in the elution buffer causes the occupancy of the vacant site of the heme iron in AoCld. The UV–visible spectrum of the Nickel Sepharose purified AoCld showed a sharp Soret peak at 413 nm with a high absorptivity as well as α and β bands at 562 and 535 nm, which indicates a 6-coordinated low spin (LS) ferric heme of the imidazole complex (Fig. S2a). Desalting (HiTrap Desalting and PD10) steps removed bound imidazole and the Soret peak has blue-shifted with a decreased absorptivity at 406 nm. Although a significant fraction of the bound imidazole was removed by the desalting, still an imidazole bound AoCld fraction remained. EPR spectroscopy showed that only 30% of the imidazole was removed at this stage, as determined by the characteristic LS signal of the imidazole adduct (gz,y,x = 2.96, 2.25, 1.51). This is consistent with the high affinity of AoCld for imidazole, for which a KD = 8 μM has been reported [4].
To further clean the enzyme, AoCld was dialyzed extensively and the Soret band blue-shifted from 406 to 391 nm, which is comparable to the Soret previously reported for native AoCld [4] (Figure S2a). During the whole process of purification, the Rz ratio A391nm/A278nm improved after each step (Table S2) [22]. AoCld was purified with homogeneity of > 90% as inferred from SDS-PAGE. SDS-PAGE analysis revealed the single band presence of 30 kDa (Fig S2b).
A color change of chlorite dismutase upon ligand binding
It was observed that AoCld switches its characteristic red color to green after buffer exchange from 100 mM KPi pH 7.0 to 100 mM sodium acetate buffer pH 5.0 (Fig. S1). The observed color change of AoCld in acetate buffer was reversible after subsequent buffer exchange to 100 mM KPi pH 7.0 (Fig. 2). The reversibility of the spectroscopic change excludes a covalent modification of the heme.
The red to green color change, due to the binding of acetate can be understood by examining the UV–visible spectrum. The Soret band red-shifted from 391 nm for the five-coordinate HS ferric enzyme to 404 nm for the six-coordinate ferric acetate complex. The intensity of the Soret peak increased from 99.6 to 141 mM−1 cm−1. In the region from 450–700 nm, several changes are observed (Table 1). The CT2 and Q bands blue-shifted from 510 to 502 nm, and CT1 blue-shifted from 647 to 626 nm, with a concomitant twofold increase in intensity (hyperchromicity) (Fig. 3a, b). We attribute the green color to the blue-shifted CT and Q bands. The CT bands represent charge transfer transitions from porphyrin π a2u → dxz,yz (Fe), which are positioned for HS heme proteins between 450–470 nm for CT2 and 600–650 for CT1.
UV–visible spectra of AoCld-acetate and fluoride adducts. 10 µM AoCld solutions were recorded in 1 mL quartz cuvette with 1.0 cm path length. a, bAoCld (red solid line) and AoCld-acetate adduct (green dotted line); c, dAoCld (red solid line) and AoCld-fluoride adduct (green dotted line); CT bands of acetate bound AoCld. Q bands indicated by the dashed grey box in a, c are enlarged for visual clarity in b, d
Dimeric Cld from Klebsiella pneumonia has been reported to bind fluoride (F−) resulting in a UV–visible spectrum dominated by a Soret at 404 nm and a blue-shifted CT1 band at 611 nm [15]. The F− adduct of pentameric Cld from Dechloromonas aromatica (DaCld) has similar spectral characteristics: a Soret band at 403 nm, and CT1 at 612 nm (Table 1) [10]. AoCld also binds F−, which is characterized by a Soret at 400 nm and a CT1 band at 613 nm (Fig. 3c, d and Table 1). The fluoride adduct of AoCld is green-colored, just like the acetate adduct.
Direct binding of acetate and fluoride to the heme iron of AoCld
The EPR spectrum of the resting state AoCld at pH 7.0 is characterized by two HS signals (Fig. 4a and Table 2) [5]. Both signals represent ms = ± 1/2 ground state doublets of S = 5/2 systems, designated as ‘narrow’ and ‘broad’ according to the rhombicity E/D. The narrow signal corresponds to an E/D = 0.01–0.02 and the broad signal to an E/D = 0.03–0.04. The same signals with different ratios have been found for Cld isolated from Azospira oryzae strain GR-1 [4], as well as recombinantly produced AoCld, DaCld and in Clds from Ideonella dechloratans, Magnetospirillum sp. [5, 7, 23].
EPR spectra of AoCld without and with several ligands. aAoCld 75 μM monomer concentration in 100 mM KPi pH 7.0. bAoCld-fluoride adduct in 100 mM KPi pH 7.0 containing 250 mM NaF showing the 19F hyperfine splitting. cAoCld-acetate adduct in 100 mM acetate pH 5.0 experimental spectrum and d simulation of the AoCld-acetate spectrum. Simulation parameters: two components: main component 71% gz,y, x = 5.98, 5.70, 1.985; Wzyx = 2.7, 6.0, 3.0 mT; minor component 29% gz,y, x = 6.36, 5.38, 1.985; Wzyx = 1.5, 2.0, 3.0 mT. eAoCld-Imidazole adduct in 100 mM KPi pH 7.0 containing 1 mM imidazole. EPR conditions: Microwave frequency, 9.405 GHz; Microwave power, 20 mW; Modulation frequency, 100 kHz; Modulation amplitude, 1.0 mT; Temperature, 20 K. The spectra were normalized for the maximal signal amplitude
The EPR spectrum of the acetate adduct of AoCld is characteristic of a HS ferric species. The spectrum could be simulated assuming two components in a 71 and 29% ratio (Fig. 4c, d). The primary component is more axial than the two species in resting-state AoCld. The minor component is similar to the ‘narrow’ signal of the resting state enzyme and may represent unbound enzyme (Table 2). To our knowledge, this is the second report of the EPR spectrum of an acetate adduct of a heme enzyme.
The EPR spectrum of the fluoride adduct of AoCld shows that complex is HS ferric (Fig. 4b). The splitting of the HS signal of the fluoride adduct at gx provides evidence of F− binding directly to the heme iron. The splitting is due to hyperfine interaction with the 19F, characterized by a nuclear spin I = 1/2 and a hyperfine coupling constant A(19F) = 44 Gauss. For the myeloperoxidase fluoride adduct A(19F) = 35 Gauss has been reported [24]. For myoglobin and hemoglobin values of 42.9 and 44.4 Gauss have been reported, respectively [25].
Binding of the established strong field ligand imidazole resulted in a LS ferric signal characterized by g-values gzyx = 2.96, 2.25, 1.51 (Table 2, Fig. 4e). This signal is also present as a minor component in the resting state enzyme (Fig. 4a), due to trace contamination of imidazole after the purification procedure.
The EPR shows spectral changes for the acetate and fluoride adducts indicating ligand binding to the heme iron. In the case of acetate and fluoride the heme iron remains HS, showing that these ligands are weak field ligands (Fig. 4).
Dissociation constant of AoCld-Acetate
Based on the UV–visible change of the Soret band upon ligand binding, the dissociation constants were determined under different conditions using equilibrium binding titrations (Table 1, Fig. 5 and Fig. S4–S6). The apparent dissociation constant of AoCld for acetate is KD,app = 19.1 ± 0.3 mM (n = 3) at pH 5.8 and 34.4 ± 5.6 mM (n = 5) at pH 5.0. Given the pKa = 4.76 of acetate the amount of ionized acetate anion, which is the most likely ligand to AoCld, is different at pH 5.0 and 5.8 [26]. Correction of the KD,app values for the pKa of acetate using the Henderson–Hasselbalch equation results in a KD = 17.5 ± 0.5 mM at pH 5.8 and 21.9 ± 3.6 mM at pH 5.0 (calculation given in the Supplementary information). Clearly, the pH dependence of the apparent dissociation constants was consistent with the relative concentration of the ionized form of acetate. Preliminary data on acetate binding to DaCld has been reported, and a KD = 36 ± 2 mM was found at pH 6.5 [27].
Representative equilibrium binding titrations AoCld with acetate, fluoride and imidazole. a, b titration of 8.40 μM AoCld in 100 mM citrate–phosphate pH 5.0 with 1 M sodium acetate; c, d titration of 8.97 μM AoCld in 100 mM KPi pH 7.0 with 1 M sodium fluoride; e, f titration of 8.89 μM AoCld in 100 mM Tris pH 8.0 with 2 mM imidazole. The first trace of the titration is in blue, the final trace is in red
The dissociation constant of AoCld for fluoride KD = 15.6 ± 1.9 mM (n = 2) at pH 7.0, which is close to the reported dissociation constant for DaCld (Table 1). As a reference, the binding of a well-established high-affinity ligand imidazole to AoCld was measured at different pH values (Fig. S6). The dissociation was KD = 74 ± 2 μM (n = 1) at pH 5.0, KD = 47.3 ± 5.1 μM (n = 3) at pH 5.8, KD = 10.8 ± 2.6 μM (n = 3) at pH 7.0 and KD = 10.5 ± 2.4 μM (n = 3) at pH 8.0. This is in excellent agreement with the reported KD = 8.8 ± 0.2 μM at pH 7.0 for native AoCld and KD = 9.6 ± 0.3 μM at pH 7.0 for native DaCld (Table 1) [4, 28]. The KD of Cld for imidazole is the lowest at pH 7.0 and 8.0 (Fig. S6k), which suggest that the affinity for the neutral imidazole is higher than for the imidazolium cation.
Based on the KD the ΔG’0 for ligand binding can be calculated, which corresponds to –10.0 ± 0.06 kJ mol−1 for acetate (pH 5.8), − 10.3 ± 0.3 kJ mol−1 for fluoride (pH 7.0) and -28.3 ± 0.6 kJ mol−1 for imidazole (pH 7.0).
Effect of acetate binding on AoCld activity and stability
With the exchange of buffer from acetate pH 5.0 to KPi pH 7.0, 60% specific activity was recovered after 15 min incubation in acetate buffer, pH 5.0. Acetate binding was found to affect AoCld activity. The observed AoCld specific activity in 100 mM KPi pH 7.0, 100 mM acetate buffer pH 5.0, and 100 mM citrate phosphate buffer pH 5.0 was 8.52·103 U/mg, 4.96·103 U/mg and 8.27·103 U/mg, respectively. It was also observed that acetate buffer pH 5.0 affected the enzyme stability. Prolonged incubation (2 h) resulted in a 90% activity loss (Fig. S3).
Discussion
Acetate binds directly to the heme iron in AoCld
Acetic acid has a pKa = 4.76 [26]. At pH 5.8 approximately 93% of the acid is present in its ionized form as calculated using HySS [29]. We conclude that acetate is an anionic ligand for AoCld. The blue-shift of the CT1 band in the UV–visible spectrum, similar to the binding of fluoride, is evidence of acetate binding directly to the heme iron. Furthermore, the EPR spectrum of the acetate adduct showed a novel HS signal that is significantly more axial than the HS signals observed for the unliganded enzyme. The binding of acetate as a direct ligand to Fe is rare and has been observed, to our knowledge, only for one other heme protein: leghemoglobin. Leghemoglobin is a high-affinity oxygen-binding protein, related to myoglobin, produced by legume plants in root nodules as part of the symbiotic relationship with nitrogen-fixing bacteria. Wittenberg et al. showed that acetate strongly reduced the rate of ferric iron reduction by sodium dithionite [30]. The UV–visible and EPR spectral properties of the acetate complex provided evidence for a high-spin complex associated with a subtle blueshift of the CT1 band from 627 to 622 nm [31]. Leghemoglobin has a distal histidine that forms a hydrogen bond with oxygen in oxyleghemoglobin [32]. It is likely that the distal histidine is also involved in binding of acetate to this protein in the ferric state.
Acetate binding to a distal site in HRP has been reported [33, 34]. The KD of HRP for acetate is > 300 mM. A small redshift of the CT1 band was observed. The Soret band did not change, but an increased asymmetry of the peak was observed. EPR spectroscopy of the HRP-acetate adduct showed the complex is HS with g-values that are similar to the unliganded enzyme. The structure of HRP-acetate showed that acetate was bound parallel to the heme plane with both carboxylate oxygens H-bonding to the proximal Arg [34]. The minimal distance between the oxygen atoms of acetate and the iron was 3.6 Å, which showed that there is no direct coordination of acetate to the iron in HRP.
Why is the AoCld-acetate complex green?
Both the Soret and Q bands represent π → π* transitions of the porphyrin ring system. The charge transfer bands from 500–600 nm represent π a2u → dxz,yz (Fe) based on a study with myoglobin single crystals [35]. AoCld in KPi pH 7.0 has the characteristic UV–visible spectrum of a 5-coordinate HS ferric heme enzyme, with a broad Soret band at 391 nm, broad overlapping Q bands at 510 nm and a clear CT1 band at 647 nm. The green color of AoCld in acetate buffer is caused by the blueshift of the CT1 band due to direct binding of acetate to the heme iron in the enzyme, in a similar way as for fluoride which also forms a green-colored complex with AoCld. We conclude that for any weak field ligand binding to HS ferric Cld, which is anionic or at least a good Lewis base a similar spectral change is expected and hence the resulting complex will also have a green color.
The interaction of the p-orbitals of the ligand with the d-orbitals of the heme iron causes an increase in the energy of the metal d-orbitals, which is responsible for the blueshift of the CT1 band. The stronger this ligand to metal interaction, the stronger the blueshift. Hydrogen bonding of a distal amino acid residue with the ligand influences the strength of the interaction. A hydrogen bond donor would lead to a stronger interaction between the ligand and the metal, leading to a stronger blueshift of the CT1 band. A hydrogen bond acceptor would lead to a redshift of the CT1 band. The CT1 band of the fluoride adduct has been considered to be a sensitive probe for the strength of the H-bonding donation of the distal amino acid residues to the F− ion. The CT1 maximum has been considered to be sensitive to the distal ligand interaction [36]. The CT1 band is sensitive to the polarity of the axial ligand as well. For example, a blueshift of 8 nm has been observed for an exchange of the axial His to a Glu, i.e. exchanging for a more negative ligand, in the 5-coordinate HS ferric cytochrome c peroxidase [37].
AoCld-acetate complex in comparison to other green heme enzymes
Spectroscopic changes that give heme proteins a green color have been reported with different causes. Oxidation of heme to verdoheme catalyzed by heme oxygenase as part of the oxidative heme degradation pathway results in such a color change [38]. The UV–visible spectrum of verdoheme is characterized by a Soret band with a reduced intensity at 405 nm and a broad absorbance band from 650–700 nm. Additionally, naturally occurring green heme proteins have been reported, the most prominent one being myeloperoxidase. It is interesting that myeloperoxidase catalyzes the formation of hypochlorite from chloride and hydrogen peroxide, which is in some ways reminiscent of the reversed reaction of chlorite dismutase. The UV–visible spectrum of myeloperoxidase is characterized by a Soret band at 428 nm and α band at 570 nm, which has been attributed to a red-shifted α band [39]. The characteristic spectral properties of myeloperoxidase are due to three covalent heme-protein bonds, which cause considerable bending of the heme plane leading to the out-of-plane location of the iron [40]. Other naturally occurring green heme proteins have also been isolated from microorganisms such as the Neurospora crassa catalase and the green heme protein from the purple acid bacterium Halochromatium salexigens [41, 42]. Finally, the seminal intermediate in many heme enzymes, compound I, an oxoferryl porphyrin π cation radical, is green-colored, e.g. in Cytochrome P450 and HRP [43, 44].
Why is the binding of acetate to AoCld similar to F− binding and not Cl− binding?
The acetate complex of Cld could offer an interesting structural model for the Michaelis complex of the enzyme. In chlorite, the O–Cl–O bond angle is 111°, whereas the O–C–O bond angle in acetate is 129° [45]. The Cl–O bond length in chlorite is 156 pm, whereas the C–O bond length in acetate is 127 pm. The required hydrogen bonding interaction between bound acetate and Arg183 could be similar to the Michaelis complex structure.
The higher affinity of Cld for F− as compared to Cl− is related to the Lewis base hardness, F− being a harder acid than Cl− and binding better to the borderline hard acid Fe3+. F− is a weak field ligand in the context of ligand field theory, as it is a π donor ligand leading to a small Δo. The spin state data in Table 2 are consistent with the spectrochemical series. Acetate as a ligand in inorganic chemistry has been reported to be close to the water in the spectrochemical series [46, 47]. This is consistent with the HS state iron in the Cld-Acetate adduct.
The blueshift and increase of intensity of the CT1 band for the acetate and fluoride complexes are related to the π donor character of both ligands. The acetate and fluoride complexes are likely stabilized by hydrogen bonding interaction with Arg183. Mutagenesis of Arg183 to Ala results in the complete loss of affinity for fluoride and acetate, but not for imidazole (Fig. S7).
Interestingly Cl− is not a suitable ligand for Cld, even though it is a product of the reaction. Cl− is a much weaker and softer base as compared to F−. Cl− binding to Cld has been reported before, but for DaCld no affinity could be measured, for the dimeric Klebsiella pneumoniae Cld cooperative binding with a KD of 1.4 mM and a Hill coefficient of 2.3 was found [15], and only a very weak affinity was observed for AoCld as no saturation was observed up to 2.5 M (not shown).
The distal Arg is important for high-affinity binding of F−, CN− and N3−, but not imidazole in DaCld [16]. This strengthens the idea that the binding of the anionic ligands involves hydrogen bonding with the distal Arg in a so-called closed conformation. We have shown that acetate binding also likely involves hydrogen bonding to Arg183.
Physiological relevance of the binding of acetate to AoCld
There are not many reports on the absolute intracellular acetate concentration in bacteria. The original organism from which this Cld was isolated, Azospira oryzae GR-1, grows anaerobically on a mineral medium with 2 g L−1 (24 mM) sodium acetate as sole carbon and electron source and perchlorate as electron acceptor [48]. The expression level of chlorite dismutase in A. oryzae GR-1 under these conditions has been estimated to be high, circa 7% of the total protein. Native chlorite dismutase has a leader sequence for periplasmic targeting. It is not unlikely that the periplasmic acetate concentration is similar to the extracellular acetate concentration and, therefore, circa 50% of natively expressed Cld may be acetate bound. The Ki and mechanism of inhibition by acetate was not determined, so we cannot estimate the precise effect on the activity in the presence of a certain local chlorite concentration. However, in conclusion, it is possible that the acetate effect we report has physiological relevance and may offer a link between the central carbon metabolism and the anaerobic respiratory pathway in this organism.
Here we present that AoCld binds acetate directly to iron in the ferric state. The resulting high spin ferric complex is characterized by a marked blueshift and hyperchromicity of the CT1 band, which gives the complex a green color. Similar effects can be expected for any ligand binding directly to the heme iron forming a high-spin ferric complex, as is indeed the case for fluoride.
Change history
29 May 2020
In the original article published, in the gy��value (column) of the H2O/OH���species (row) of Table 2 was mistakenly given as ���1.18��� and the correct value is ���2.18���.
Abbreviations
- Ac:
-
Acetate
- AoCld:
-
Azospira oryzae Chlorite dismutase
- Cld:
-
Chlorite dismutase
- DaCld:
-
Dechloromonas aromatica Chlorite dismutase
- EPR:
-
Electron paramagnetic resonance
- HS:
-
High spin
- HRP:
-
Horseradish peroxidase
- Im:
-
Imidazole
- IMAC:
-
Immobilized metal ion affinity chromatography
- IPTG:
-
Isopropyl ß-d-1-thiogalactopyranoside
- KPi:
-
Potassium phosphate buffer
- LS:
-
Low spin
- Rz:
-
Reinheitszahl
References
Coates JD, Achenbach LA (2004) Nat Rev Microbiol 2:569–580
Ginkel CG, Rikken GB, Kroon AGM, Kengen SWM (1996) Arch Microbiol 166:321–326
Rutherford AW (1989) Trends Biochem Sci 14:227–232
Hagedoorn PL, De Geus DC, Hagen WR (2002) Eur J Biochem 269:4905–4911
Freire DM, Rivas MG, Dias AM, Lopes AT, Costa C, Santos-Silva T, Van Doorslaer S, González PJ (2015) J Inorg Biochem 151:1–9
De Geus DC, Thomassen EAJ, Hagedoorn PL, Pannu NS, Van Duin E, Abrahams JP (2009) J Mol Biol 387:192–206
Stenklo K, Thorell HD, Bergius H, Aasa R, Nilsson T (2001) J Biol Inorg Chem 6:601–607
Mlynek G, Sjöblom B, Kostan J, Füreder S, Maixner F, Gysel K, Furtmüller PG, Obinger C, Wagner M, Daims H, Djinović-Carugo K (2011) J Bacteriol 193:2408–2417
Goblirsch B, Kurker RC, Streit BR, Wilmot CM, DuBois JL (2011) J Mol Biol 408:379–398
Hofbauer S, Hagmüller A, Schaffner I, Mlynek G, Krutzler M, Stadlmayr G, Pirker KF, Obinger C, Daims H, Djinović-Carugo K, Furtmüller PG (2015) Arch Biochem Biophys 574:36–48
Hofbauer S, Howes BD, Flego N, Pirker KF, Schaffner I, Mlynek G, Djinović-Carugo K, Furtmüller PG, Smulevich G, Obinger C (2016) Biosci Rep 36:e00312
Afshar S, Johnson E, Vries S, Schröder I (2001) J Bacteriol 183:5491–5495
Yoshimatsu K, Sakurai T, Fujiwara T (2000) FEBS Lett 470:216–220
Geeraerts Z, Celis AI, Mayfield JA, Lorenz M, Rodgers KR, DuBois JL, Lukat-Rodgers GS (2018) Biochemistry 57:1501–1516
Geeraerts Z, Rodgers KR, DuBois JL, Lukat-Rodgers GS (2017) Biochemistry 56:4509–4524
Blanc B, Mayfield JA, McDonald CA, Lukat-Rodgers GS, Rodgers KR, DuBois JL (2012) Biochemistry 51:1895–1910
Streit BR, Blanc B, Lukat-Rodgers GS, Rodgers KR, DuBois JL (2010) J Am Chem Soc 132:5711–5724
de Geus DC, Thomassen EAJ, van der Feltz CL, Abrahams JP (2008) Acta Cryst F 64:730–732
Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC (1985) Anal Biochem 150:76–85
Salmeen I, Palmer G (1968) J Chem Phys 48:2049–2052
Lundin A, Aasa R (1972) J Magn Reson 8:70–73
Hofbauer S, Schaffner I, Furtmüller PG, Obinger C (2014) Biotechnol J 9:461–473
Lee AQ, Streit BR, Zdilla MJ, Abu-Omar MM, DuBois JL (2008) Proc Natl Acad Sci 105:15654–15659
Wever R, Bakkenist ARJ (1980) Biochim Biophys Acta Enzymol 612:178–184
Morimoto H, Kotani M (1966) Biochim Biophys Acta Biophys Photosyn 126:176–178
Haynes WM, Lide DR, Bruno TJ (2016) CRC handbook of chemistry and physics: a ready-reference book of chemical and physical data. CRC Press, Boca Raton
Goblirsch BR, Streit BR, DuBois JL, Wilmot CM (2010) J Biol Inorg Chem 15:879–888
Blanc B, Rodgers KR, Lukat-Rodgers GS, DuBois JL (2013) Dalton Trans 42:3156–3169
Alderighi L, Gans P, Ienco A, Peters D, Sabatini A, Vacca A (1999) Coord Chem Rev 184:311–318
Wittenberg BA, Wittenberg JB, Appleby CA (1973) J Biol Chem 248:3178–3182
Appleby CA, Blumberg WE, Peisach J, Wittenberg BA, Wittenberg JB (1976) J Biol Chem 251:6090–6096
Harutyunyan EH, Safonova TN, Kuranova IP, Popov AN, Teplyakov AV, Obmolova GV, Rusakov AA, Vainshtein BK, Dodson GG, Wilson JC, Perutz MF (1995) J Mol Biol 251:104–115
Araiso T, Dunford HB (1981) J Biol Chem 256:10099–10104
Carlsson GH, Nicholls P, Svistunenko D, Berglund GI, Hajdu J (2005) Biochemistry 44:635–642
Eaton WA, Hochstrasser RM (1968) J Chem Phys 49:985–995
Smulevich G, Howes BD, Droghetti E (2016) In: Dunford B, Raven EL (eds) Heme peroxidases. The Royal Society of Chemistry, Croydon, pp 61–91
Smulevich G, Neri F, Willemsen O, Choudhury K, Marzocchi MP, Poulos TL (1995) Biochemistry 34:13485–13490
Wilks A, Heinzl G (2014) Arch Biochem Biophys 544:87–95
Wever R, Plat H (1981) Biochim Biophys Acta Enzymol 661:235–239
Zederbauer M, Furtmüller PG, Brogioni S, Jakopitsch C, Smulevich G, Obinger C (2007) Nat Prod Rep 24:571–584
Jacob GS, Orme-Johnson WH (1979) Biochemistry 18:2975–2980
Van Driessche G, Devreese B, Fitch JC, Meyer TE, Cusanovich MA, Van Beeumen JJ (2006) FEBS J 273:2801–2811
Rittle J, Green MT (2010) Science 330:933–937
Blumberg WE, Peisach J, Wittenberg BA, Wittenberg JB (1968) J Biol Chem 243:1854–1862
Greenwood NN, Earnshaw A (2006) Chemistry of the elements. Butterworth-Heinemann, Oxford
Edwards DA, Hayward RN (1968) Can J Chem 46:3443–3446
van Leeuwen PWNM, Groeneveld WL (1968) Recl Trav Chim Pays-Bas 87:86–96
Rikken GB, Kroon AGM, Ginkel CG (1996) Appl Microbiol Biotechnol 45:420–426
The PyMOL molecular graphics system, Schrödinger. https://www.pymol.org/
Acknowledgements
This research was supported by Grant NWO–CW 711.014.006 from the Council for Chemical Sciences of The Netherlands Organization for Scientific Research.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mahor, D., Püschmann, J., van den Haak, M. et al. A traffic light enzyme: acetate binding reversibly switches chlorite dismutase from a red- to a green-colored heme protein. J Biol Inorg Chem 25, 609–620 (2020). https://doi.org/10.1007/s00775-020-01784-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00775-020-01784-1