Abstract
The heterotrimeric electron-bifurcating [FeFe] hydrogenase (HydABC) from Thermotoga maritima (Tm) couples the endergonic reduction of protons (H+) by dihydronicotinamide adenine dinucleotide (NADH) (∆G0 ≈ 18 kJ mol−1) to the exergonic reduction of H+ by reduced ferredoxin (Fdred) (∆G0 ≈ − 16 kJ mol−1). The specific mechanism by which HydABC functions is not understood. In the current study, we describe the biochemical and spectroscopic characterization of TmHydABC recombinantly produced in Escherichia coli and artificially maturated with a synthetic diiron cofactor. We found that TmHydABC catalyzed the hydrogen (H2)-dependent reduction of nicotinamide adenine dinucleotide (NAD+) in the presence of oxidized ferredoxin (Fdox) at a rate of ≈17 μmol NADH min−1 mg−1. Our data suggest that only one flavin is present in the enzyme and is not likely to be the site of electron bifurcation. FTIR and EPR spectroscopy, as well as FTIR spectroelectrochemistry, demonstrated that the active site for H2 conversion, the H-cluster, in TmHydABC behaves essentially the same as in prototypical [FeFe] hydrogenases, and is most likely also not the site of electron bifurcation. The implications of these results are discussed with respect to the current hypotheses on the electron bifurcation mechanism of [FeFe] hydrogenases. Overall, the results provide insight into the electron-bifurcating mechanism and present a well-defined system for further investigations of this fascinating class of [FeFe] hydrogenases.
Graphic abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
[FeFe] hydrogenases catalyze the reversible interconversion of protons (H+) and electrons to hydrogen (H2) at very high rates, with negligible energy waste [1,2,3]. Their active center, the H-cluster, consists of a unique [2Fe] cluster ([2Fe]H) tethered covalently by a cysteine thiolate to a standard [4Fe–4S] cluster ([4Fe–4S]H) (Fig. 1a) [4, 5]. The Fe ions in the [2Fe]H subsite are coordinated by CO and CN− ligands and also by a bidentate 2-azapropane 1,3-dithiolate (ADT) ligand that bridges the two irons of [2Fe]H (Fig. 1a). The H-cluster is buried inside a highly optimized protein scaffold, which tunes its catalytic efficiency and provides pathways for the transport of protons, electrons, and gases (the substrate H2 as well as inhibitors such as CO and O2) [6].
a Ball and stick representation of the structure of the H-cluster. The figure was created in Pymol using PDB file 4XDC. b Schematic diagram showing the three subunits of TmHydABC. The cofactors bound to the protein were predicted from amino acid sequence analysis [26]. The arrangement of subunits and the designated reactions occurring at each subunit are based on the proposition by Buckel and Thauer [23]
During H2 conversion, the H-cluster passes through several redox states [1]. In the active oxidized state (Hox), the [2Fe]H subcluster is in a mixed valent [Fe(I)Fe(II)] state, and the [4Fe–4S]H subsite is oxidized (2 +). One-electron reduction of the [4Fe–4S]H subsite forms the Hred state [7]. Protonation of the ADT in the Hred state is coupled to electron transfer from the reduced [4Fe–4S]H to the [2Fe] cluster, yielding the HredH+ state [8]. A second electron reduction forms the most reduced state, HsredH+ ([4Fe–4S]1+–[Fe(I)Fe(I)]) [9]. Rearrangement of the HsredH+ state gives the Hhyd state, where a terminal hydride is bound to the [2Fe] subsite with an Fe(II)Fe(II) configuration and [4Fe–4S]H remains reduced [10, 11]. Protonation of Hhyd leads to H2 production and regeneration of the Hox state.
Based on amino acid sequence phylogeny, the [FeFe] hydrogenases have been classified into three major groups: (I) prototypical and electron-bifurcating; (II) ancestral; and (III) sensory [12]. The physiological roles of the prototypical [FeFe] hydrogenases are far better understood than the other classes. While the prototypical [FeFe] hydrogenases use ferredoxin or cytochrome as their sole redox partner, the electron-bifurcating [FeFe] hydrogenases use both ferredoxin (Fd) and dihydronicotinamide adenine dinucleotide (NADH) simultaneously during H2 evolution [13,14,15,16].
Electron bifurcation is a process, whereby an endergonic redox reaction and an exergonic redox reaction are directly coupled, and is an alternative energy conservation mechanism to the well-known chemiosmotic coupling mechanism [16, 17]. First described by Peter Mitchell in the Q-cycle of mitochondrial complex III [18], it was only about a decade ago that the involvement of electron bifurcation in the metabolism of anaerobic microorganisms was discovered [19]. Subsequent investigations showed that in a variety of reactions in anaerobic metabolism, electron bifurcation is involved [20], among them: coupling of the endergonic oxidation of NADH (E0′ = − 320 mV) to the exergonic oxidation of reduced ferredoxin (Fdred) (E0′ ≈ − 450 mV) to reduce H+ to H2 (E0′ = − 420 mV). The overall reaction is the recycling of NADH, generated by the oxidation of sugars, driven by Fdred oxidation and can be described in full as [14, 16, 21,22,23]:
This reaction is essential in many organisms as various metabolic steps only yield enough energy to reduce nicotinamide adenine dinucleotide (NAD+), but NADH is not capable of reducing protons. Thus, by coupling NADH oxidation to ferredoxin oxidation, protons can be used as a terminal electron acceptor [20, 21].
Electron-bifurcating [FeFe] hydrogenases are widely found in the genomes of anaerobic bacteria belonging to the phyla of Bacteroidetes, Firmicutes, Spirochaetes, Thermotogae, and Fusobacteria [12]. Bacteria belonging to Bacteroidetes and Firmicutes phyla are commonly found in mammalian guts, and as H2 is an abundant metabolite in the gastrointestinal tract, hydrogenases found in the above-mentioned organisms play an important role in colonic H2 metabolism [24]. Interestingly, in a recent bioinformatics study, it has been shown that a wide variety of hydrogenases found in the bacteria of the human gut belong to the electron-bifurcating class [25]. Therefore, understanding the metabolism of these organisms may also be important for medical applications.
Electron-bifurcating [FeFe] hydrogenases have been characterized from a number of organisms; however, the main focus of all these studies were their biochemical properties [14, 21, 27,28,29]. Even though the electron-bifurcating [FeFe] hydrogenase from Thermotoga maritima (Tm) has been spectroscopically characterized [26, 30], ambiguities in the understanding of its biophysical properties still persist. In this work, based on the previous efforts of recombinant expression and artificial maturation of [FeFe] hydrogenases, we have developed a recombinant system to overexpress TmHydABC in E. coli followed by its artificial maturation in vitro. This system allows the generation of high yields of pure enzyme, making it possible to perform a complete spectroscopic analysis to study in detail the electron-bifurcating mechanism of TmHydABC [FeFe] hydrogenase.
The genome of the hyperthermophilic bacterium, T. maritima, contains genes encoding a prototypical, a sensory, and an electron-bifurcating type [FeFe] hydrogenase [14, 21, 31]. Previous biochemical studies showed that the electron-bifurcating [FeFe] hydrogenase, TmHydABC, is the key enzyme responsible for H2 production in T. maritima [14]. The electron-bifurcating [FeFe] hydrogenase from T. maritima is a trimeric protein composed of TmHydA, TmHydB, and TmHydC (Fig. 1b) [26]. The largest subunit, TmHydA, harbors the H-cluster along with three additional [4Fe–4S] and two [2Fe–2S] clusters. TmHydB is the second largest subunit and consists of three [4Fe–4S] clusters, one [2Fe–2S] cluster, a flavin mononucleotide (FMN) binding domain, and an NADH-binding motif. TmHydC is the smallest of the three subunits and contains only one [2Fe–2S] cluster [32, 33].
The mechanistic study of prototypical [FeFe] hydrogenases has greatly benefited from the discovery of ‘artificial maturation’, whereby heterologously produced apo-proteins, that is the protein containing the [4Fe–4S]H sub cluster but lacking [2Fe]H, have been reconstituted using chemically synthesized precursor complexes [34, 35]. Here, we have expanded the repertoire of hydrogenases to which artificial maturation can be applied by employing it to produce the trimeric TmHydABC and also the catalytic subunit TmHydA. Subsequently, we have performed biochemical assays with the resulting hydrogenase as well as a full spectroscopic characterization of the H-cluster. The results reveal insight into the bifurcation mechanism and even more importantly provide the basis for further investigation of this unresolved phenomenon.
Materials and methods
Activity assays
Hydrogen oxidation activity of TmHydABC and TmHydA was measured by following the reduction of 1 mM benzyl viologen (at 600 nm, ε600 = 7.8 mM−1 cm−1) in 200 mM H2-saturated potassium phosphate buffer, pH 8. The reactions were performed at various temperatures (30–80 °C) by the addition of 25–50 ng protein to 1 mL of the above-mentioned reaction buffer in 1.5 mL plastic cuvettes and the change in absorbance was measured using an Ocean Optics DH-mini UV–Vis–NIR light source and a USB2000 + XR1-ES detector, operated by the SpectraSuiteTM software. The desired reaction temperatures were achieved using a temperature controlled cuvette holder (CUV-QPOD-2E-ABSKIT, Ocean Optics). All values are the average of three measurements after subtracting the value of the blank measurement. The other details are described in the figure captions.
The NAD+ and ferredoxin-dependent H2 oxidation assay of TmHydABC was performed at 70 °C by addition of 300–600 ng of the protein to 1 mL of H2 saturated 200 mM potassium phosphate buffer, pH 8. The formation of NADH (monitored at 340 nm, ε = 6.2 mM−1 cm−1) and reduction of ferredoxin (monitored at 430 nm, ε = 12 mM−1 cm−1) were determined from the change in absorbance measured using a spectrophotometer.
To determine the methyl viologen dependent hydrogen production, 250–500 ng of protein was added to 10 mM methyl viologen reduced with 100 mM sodium dithionite (NaDT) in 200 mM potassium phosphate buffer pH 8 in 2.5 mL plastic tubes with rubber stoppers. The reaction mixture was purged with argon for 5 min and incubated at the desired reaction temperature for 10 min before 0.5 mL of the headspace gas was extracted for analysis. The head space gas was then analyzed by gas chromatography. Hydrogen content was quantified by comparison with a 100% H2 standard. All values are the average of three measurements after subtracting the value of a blank measurement.
Electrochemistry
Protein film electrochemistry experiments were performed using a gas-tight three electrode setup inside an anaerobic glovebox (MBraun) filled with N2. The cell temperature was controlled by a water-jacket system. A pyrolytic graphite edge disk (0.03 cm2, Momentive Materials) was used as a working electrode and a Pt wire as a counter electrode. The reference electrode (saturated calomel electrode—SCE) was placed in a side arm (at room temperature), connected to the main cell compartment by a Luggin capillary. Electrochemical experiments were controlled by a VersaStat 4 potentiostat (Princeton Applied Research). The electrochemical cell was filled with a buffer mixture of MES, HEPES, TAPS, CHES, and sodium acetate adjusted to pH 5, 6, 7, 8, 9, and 10. Cyclic voltammetry was performed under 100% H2 with a 2000 rpm rotation rate (Princeton Applied Research model 636A). The potentials were converted to the standard hydrogen electrode (SHE) using a conversion of + 241 mV from SCE.
FTIR spectroscopy
FTIR spectroscopy was performed using home-built water-cooled sample holders accommodated in a Bruker Vertex v80 spectrometer. Samples (10 μL) were placed between two CaF2 (Korth Kristalle, Altenholz) windows, separated by a 50 μm Teflon spacer, and sealed with rubber rings. The temperature of the sample holder was maintained using a water circulator system (Huber, Offenburg). The spectrometer was equipped with a liquid nitrogen cooled fast mercury cadmium telluride photovoltaic (D317/B) detector. Spectra were collected at room temperature in double-sided, forward–backward mode with a 2 cm−1 resolution and 20 kHz scanner velocity.
FTIR spectroelectrochemistry
Spectroelectrochemical experiments were performed using a home-built electrochemical cell according to the design described by Moss et al. [1, 36]. Protein samples (1–1.5 mM) containing 0.5 mM of the redox mediators benzyl viologen (Em = − 358 mV) and methyl viologen (Em = − 449 mV) were loaded between two CaF2 (Korth Kristalle, Altenholz) windows on a gold mesh working electrode (approximately 20 μm thick) in electrical contact with a platinum counter electrode. An Ag/AgCl (1 M KCl) electrode was used as a reference and was calibrated before and after the measurement with (hydroxymethyl)ferrocene (Em = + 436 mV). In the titrations, the potential was controlled by an Autolab PGSTAT101 potentiostat controlled by the Nova software.
The measurements were performed using a Bruker IFS 66v/S FTIR spectrometer equipped with a mercury cadmium telluride photovoltaic detector in double-sided, forward–backward mode with 2 cm−1 resolution, and 20 kHz scanner velocity. The temperature (15 °C) was maintained by a water circulator (Huber). Spectra were collected after 40 min incubation time at each applied potential.
EPR spectroscopy
X-band EPR spectra were recorded on a Bruker ELEXSYS E500 CW X-band EPR spectrometer. The temperature of the samples was controlled using an Oxford Instruments ESR900 helium flow cryostat connected to an ITC503 temperature controller. The measurement parameters were: microwave frequency 9.64 GHz, time constant 81.92 ms, conversion time 81.92 ms, and modulation frequency 100 kHz. The microwave power and temperature were varied between measurements and are indicated in the figure legends.
The EPR samples (200 µL) were transferred anaerobically to 4 mm (o.d.) quartz EPR tubes and frozen in liquid nitrogen. All spectra were analyzed with home-written scripts in MATLAB. Spectral simulations were performed using the EasySpin package [37]. Spin quantification was achieved by comparison with a 1 mM CuSO4, 10 mM EDTA standard.
Results
Characterization of apo-TmHydABC and apo-TmHydA
The apo-TmHydABC and apo-TmHydA were produced recombinantly in E. coli (details of construct preparation, heterologous expression, and purification are provided in the Supplementary Information). Iron quantification of apo-TmHydABC and apo-TmHydA indicated the presence of 35 ± 2 and 19 ± 1 moles of iron per mole of protein, respectively, in good agreement with the expected values (36 for apo-TmHydABC and 20 for apo-TmHydA) based on the number of iron–sulfur (FeS) clusters, and on the previously reported data on TmHydABC and TmHydA isolated from the native organism [26]. Flavin quantification gave 0.2 ± 0.05 moles of FMN per mole of protein. Inclusion of riboflavin during protein overexpression did not enhance the FMN content. Thus, it is likely that the majority of the FMN is lost during purification, as has been demonstrated also for native TmHydABC [26].
Apo-TmHydABC and apo-TmHydA were analyzed using UV–Vis and continuous wave (CW) electron paramagnetic resonance (EPR) spectroscopy (Figure S5). In both cases, the UV–Vis spectrum showed broad bands in the range from 300 to 600 nm (Figure S5A) typical for [2Fe–2S] and [4Fe–4S] clusters. EPR spectra from apo-TmHydABC and apo-TmHydA reduced with sodium dithionite are shown in Figure S5B. The EPR spectrum of apo-TmHydABC at 40 K is similar to that observed with the reduced native protein and has g values and relaxation properties consistent with [2Fe–2S] clusters (Figure S5B, upper panel) [26]. The EPR spectrum of apo-TmHydABC at 10 K appears to be more complex than that at 40 K, with the broadening of the feature at g ≈ 2.02 and appearance of a new feature at g ≈ 1.88 (Figure S5B, upper panel). This phenomenon was also observed in the native protein and was attributed to the weak dipolar interactions between the clusters [26]. The EPR spectrum of apo-TmHydA is very similar to the apo-TmHydABC EPR spectrum at 40 K (Figure S5B, lower panel). Interestingly, at 10 K the EPR spectrum of TmHydA does not show such a large contribution from the broad feature as apo-TmHydABC. Nevertheless, it can be concluded that apo-TmHydABC and apo-TmHydA contain both [2Fe–2S] and [4Fe–4S] clusters essentially identical to that of the proteins purified from the native organism.
Activity assays with artificial partners
The apo-TmHydABC and apo-TmHydA were artificially maturated using the previously published in vitro method (see Supplementary Text 3 for details) to generate the holo-enzymes [34, 35]. Activity assays with holo-TmHydABC and holo-TmHydA in solution were performed to obtain the rates of H2 oxidation coupled to benzyl viologen reduction and H2 production coupled to methyl viologen oxidation. At 30 °C, TmHydABC and TmHydA oxidized H2 at a rate of ≈500 U mg−1 and ≈800 U mg−1, respectively (1 U = 1 µmol min−1) (Fig. 2a). The H2 production activities of TmHydABC and TmHydA at 37 °C were found to be ≈270 U mg−1 and ≈150 U mg−1, respectively (Fig. 2b). While both activities increased with temperature, a stronger effect was found for H2 oxidation (Fig. 2). Compared to TmHydABC isolated from the native organism the artificially maturated protein showed significantly higher activity [38]. This could be related to a higher conversion of the CO-inhibited state Hox–CO to the active states in the artificially maturated protein, while in the native organism, a larger fraction of the isolated protein is in the inactive Hox–CO state.
Activity assays of artificially maturated TmHydABC and TmHydA. a Temperature dependence of H2 oxidation of both proteins was measured in the range from 30 to 80 °C with benzyl viologen as the electron acceptor. b H2 production activity of TmHydABC and TmHydA was measured at 37 °C and 70 °C with reduced methyl viologen as electron donor
Catalytic activity using protein film electrochemistry
The catalytic activity of TmHydABC and TmHydA was also measured electrochemically by adsorbing the enzymes on pyrolytic graphite electrodes (Fig. 3). Due to protein desorption at increased temperatures, the maximum temperature that could be used was 35 °C. Under these conditions reasonably stable films were formed such that individual voltammograms could be measured over a range of different pH values. From these measurements, it was clear that higher current densities are achieved with TmHydA (up to − 0.9 mA cm−2 for H+ reduction) (Fig. 3b) than TmHydABC (up to − 0.045 mA cm−2 for H+ reduction) (Fig. 3a), but otherwise, the two enzymes behaved in a similar fashion showing a bias towards H+ reduction at neutral to low pH. The difference in current densities between TmHydA and TmHydABC could be related to the larger size of TmHydABC (160 kDa) compared to TmHydA (72 kDa), which may lead to higher electroactive coverage (surface concentration of the electroactive enzyme) for TmHydA, as more enzymes can fit into the same surface area. The oligomeric nature of the enzyme can also influence the electroactive coverage: TmHydABC forms a tetramer of trimers in solution (around 700 kDa), while TmHydA behaves as a monomer or dimer (Figures S2 and S3). Protein film electrochemistry reveals that for both TmHydABC and TmHydA, the H+ reduction current is strongly pH dependent, dominating over H2 oxidation at pH values below 8, while the H2 oxidation current is relatively pH independent (Fig. 3). Both enzymes show reversible electrochemical behavior with essentially no over-potential requirement in either direction (Fig. 3). The behavior of TmHydA is reminiscent of that from Clostridium pasteurianum and Clostridium acetobutylicum [FeFe] hydrogenases both of which show a bias towards H+ reduction at neutral pH [9, 39].
Cyclic voltammetry of TmHydABC (a) and TmHydA (b) adsorbed onto the surface of a pyrolytic graphite electrode. The applied potential was swept from open-circuit potential to 250 mV more positive and then to 250 mV more negative, to compare the catalytic currents at the same overpotential. The measurements were performed at 35 °C under 100% H2
Catalytic activity using physiological partners
The TmHydABC enzyme isolated from the native organism was unable to produce H2 with reduced ferredoxin as the sole electron donor [14]. However, when reduced ferredoxin and NADH were added together as electron donors, TmHydABC could evolve H2 [14]. Thus, TmHydABC acts as an electron-confurcating (reverse of bifurcation) hydrogenase, which converges electrons from two different sources to reduce protons to H2 [14, 23]. To check if the recombinantly produced and artificially maturated TmHydABC is able to perform electron bifurcation, we followed the reverse reaction, i.e., we monitored H2 oxidation coupled to reduction of NAD+ and ferredoxin. The ferredoxin (TmFd1) used for this assay is a single [4Fe–4S] cluster containing ferredoxin from T. maritima (details of its purification and characterization are provided in the Supplementary Text 4 and Figure S8). TmHydABC was not able to reduce TmFd1 in an H2-saturated assay mixture containing FMN but lacking NAD+ (indicated by the unchanged absorbance intensity at 430 nm) (Fig. 4a). As soon as NAD+ was added to the assay, a decrease in the absorbance at 430 nm was observed, indicating reduction of TmFd1 (Fig. 4a). Concomitantly, NADH production was detected by an increase of the absorbance at 340 nm (Fig. 4a). The changes in the absorbance intensities at 340 nm and 430 nm were not observed in the absence of TmHydABC, indicating that the H2-dependent reduction of NAD+ and TmFd1 is indeed catalyzed by TmHydABC.
Activity assay of TmHydABC using physiological partners. a Simultaneous reduction of TmFd1 (decrease in A430nm) and NAD+ (increase in A340nm) by TmHydABC at 70 °C under an atmosphere of 100% H2 was monitored by UV–Vis spectroscopy. To a 1 mL reaction mixture containing ≈ 680 ng TmHydABC and 50 µM FMN in 200 mM potassium phosphate (pH 8) buffer, ≈35 µM TmFd1 was added (indicated by the first arrow). To the same reaction mixture, 0.5 mM NAD+ was added (indicated by the second arrow). b Reduction of TmFd1 monitored by EPR spectroscopy. In the absence of TmHydABC, H2 did not reduce TmFd1 (trace i). In the absence of NAD+, TmHydABC could not catalyze reduction of TmFd1 from H2 (trace ii). However, when NAD+ is present, TmHydABC could reduce TmFd1 using H2 as indicated by the appearance of the typical rhombic spectrum corresponding to the reduced [4Fe–4S] cluster. The conditions of the reactions used for preparing EPR samples were the same as for the UV–Vis measurements. The EPR spectra were measured at 10 K and at 0.2 mW power
The above observation was also confirmed by EPR spectroscopy (Fig. 4b). The reaction mixture containing NAD+ and TmFd1, but lacking TmHydABC, does not show EPR signals corresponding to the reduced [4Fe–4S] cluster of TmFd1 (Fig. 4b, trace i). Similarly, no [4Fe–4S] cluster EPR signal was observed when the reaction was performed without NAD+ (Fig. 4b, trace ii). The typical rhombic feature of the reduced [4Fe–4S] cluster originating from TmFd1 was observed only when the reaction mixture contained both NAD+ and catalytic amounts of TmHydABC (Fig. 4b, trace iii). These results indicate that our holo-TmHydABC is capable of electron bifurcation in the presence of its physiological redox partners.
The specific activity of NADH production by TmHydABC in the presence of TmFd1 at 70 °C was found to be ≈16.6 U mg−1 (Table S1), which is well within the range that was obtained for other electron-bifurcating [FeFe] hydrogenases [21, 27,28,29, 40]. The stoichiometry of NADH produced vs TmFd1 reduced was found to be 1:2.01 (± 0.1), indicating that the reaction proceeds as expected according to Eq. (1) (Figure S9). Like the electron-bifurcating [FeFe] hydrogenase from Desulfovibrio fructosovorans (Hnd) [29], TmHydABC was also able to reduce NAD+ in the absence of TmFd1, however, at ≈35-fold lower rate (Table S1). The specific activity of NADH production was found to be ≈fivefold lower (3.4 ± 0.5 U mg−1) when flavin adenine dinucleotide (FAD) was added to the assay buffer instead of FMN (Table S1). A similar observation was also made in the case of natively isolated TmHydABC that showed approximately 50% activity when FAD was added instead of FMN [14]. It is interesting to note that even in the absence of any FMN or FAD artificially maturated TmHydABC could form NADH at ≈fourfold lower rate (4.7 ± 0.6 U mg−1) than when excess FMN was added to the assay mixture (Table S1, Figure S10). This residual activity appears to arise from the fraction of TmHydABC molecules that retained the FMN cofactor (0.2 ± 0.05 moles of FMN per mole of TmHydABC). From this observation, it can be concluded that the flavin cofactor required for the optimal bifurcation activity of TmHydABC is FMN and not FAD. Furthermore, this observation supports the idea that only one FMN per HydABC trimer is required for bifurcation and that a second flavin is highly unlikely to be present. Since the addition of excess FMN leads to an ≈fourfold increase in rate, this indicates that the FMN content has likewise increased ≈fourfold (from ≈ 0.2 FMN per HydABC to ≈ 0.8). If there were two sites, excess FMN would be expected to increase the FMN content ≈tenfold, leading to a greater than tenfold increase in the rate. This finding has important implications for the bifurcation mechanism, which are discussed below.
Spectroscopic characterization of the holo-protein
The CO and CN− ligands coordinated to the iron atoms in the H-cluster have characteristic stretching vibrations in the range 2150–1750 cm−1, where the characteristic amide vibrations from the protein backbone are absent. Since the stretching vibrations of these ligands are sensitive to changes in oxidation state of the metal center, Fourier transform infrared (FTIR) spectroscopy is an effective tool to study the redox intermediates of the H-cluster. The characteristic FTIR bands for the various redox states of some well-characterized [FeFe] hydrogenases are presented in Table S2.
EPR spectroscopy is an additional tool providing information about the paramagnetic species present in the various redox states of the hydrogenase. Characteristic g values for the redox states of some well-characterized hydrogenases are presented in Table S3.
Characterization of the oxidized state
After artificial maturation, both TmHydABC and TmHydA were obtained in a mixture of oxidation states (Figure S6). Various redox agents were used to enrich the individual oxidation states. Figure 5 shows the FTIR and EPR spectra of TmHydABC and TmHydA after oxidizing the proteins with NAD+ and thionine, respectively. In both cases, the FTIR spectra (Fig. 5a) are characterized by five major FTIR bands (2090, 2076, 1964, 1939, and 1802 cm−1). The positions of these bands are comparable to those reported for the Hox state of TmHydABC isolated from the native organism [30] and also to those reported for other [FeFe] hydrogenases (Table S2). It is, therefore, reasonable to assume that the H-cluster of both TmHydABC and TmHydA are mostly in the Hox state under the applied oxidizing conditions.
FTIR and EPR spectra of artificially maturated TmHydABC and TmHydA under oxidizing conditions. a Top panel shows the FTIR spectrum of 400 µM TmHydABC oxidized with 20 mM NAD+ and the lower panel shows the FTIR spectrum of 400 µM TmHydA oxidized with 1 mM thionine. Both samples were in 0.1 M Tris–HCl pH 8 buffer, 0.15 M NaCl and the spectra were measured at room temperature. The peaks belonging to the Hox state are shaded in red and those belonging to the Hox–CO state are shaded in blue. b CW X-band EPR spectra of the oxidized TmHydABC and TmHydA samples at 20 K, 0.1 mW microwave power. The samples were prepared in the same way as for the FTIR measurements. The experimental spectra are overlaid with spectral simulations (dotted magenta lines) and the component spectra are shown underneath. The red trace (Component 1) corresponds to the Hox state, the blue trace (Component 2) corresponds to the Hox–CO state and the gray trace (Component 3) corresponds to one of the reduced F-clusters
In the Hox state, both irons in the [2Fe]H subsite are in a low spin configuration leading to an S = 1/2 ground state for Hox. In this electronic configuration, the H-cluster shows a rhombic EPR spectrum with two g values above 2.0. Figure 5b shows the CW X-band EPR spectra of oxidized TmHydABC and TmHydA. The experimental spectra of both TmHydABC and TmHydA were simulated with three components (details of the EPR simulations are given in Table S4). For both TmHydABC and TmHydA, Component 1 is a rhombic EPR spectrum with almost identical g values; 2.102, 2.044, and 1.998 for TmHydABC and 2.103, 2.045, and 1.999 for TmHydA (Fig. 5b). These g values are in good agreement with the g values of the Hox state of other [FeFe] hydrogenases (Table S3). Thus, the Hox state is also observed for TmHydABC and TmHydA by EPR after oxidative treatment. The additional Component 2 can be assigned to the Hox–CO state, and indications of its presence were also observed in the FTIR spectra (Fig. 5a, peaks shaded in blue). The third component with g values of [2.013, 1.950, 1.917] for TmHydABC and [2.005, 1.949, 1.918] for TmHydA (Table S4) can be assigned to reduced FeS clusters based on the similarity to the spectra of reduced apo-TmHydABC and apo-TmHydA (Figure S5B). When the EPR spectrum of the NAD+ oxidized TmHydABC sample was measured at higher temperature (40 K), signals corresponding to Component 3 could still be observed, which indicated that this signal is originating from a slowly relaxing [2Fe–2S] cluster (Figure S11). EPR spin quantitation on the oxidized holo-TmHydABC sample gave a spin content of ≈ 0.6 spins/molecule of protein for the Hox component and ≈ 0.1 spins/molecule of protein for the Hox–CO component. Therefore, the total spin content of the H-cluster is approximately 0.7 spins/molecule of protein. In the case of native TmHydABC the spin content originating from the H-cluster was found to be ≈0.1 spin/molecule of protein [26], indicating a larger proportion of intact H-clusters in the TmHydABC sample prepared by our method, which could also explain the higher activity found in solution compared with that reported for the native enzyme [26].
Characterization of the CO-inhibited state
CO is a well-known competitive inhibitor for [FeFe] hydrogenases by occupying the open coordination site of the H-cluster to form the Hox–CO state, a well-defined state showing characteristic EPR and FTIR signals [1, 41, 42]. FTIR and EPR spectra of the Hox–CO state of TmHydABC and TmHydA are shown in Fig. 6.
FTIR and EPR spectra of TmHydABC and TmHydA after CO inhibition. a FTIR spectra of CO-inhibited (Hox–CO) artificially maturated TmHydABC and TmHydA. The ‘as-isolated’ protein samples (≈ 400 µM) were purged for 20 min with 100% CO, followed by incubation for an additional 60 min. Spectra were measured at room temperature. The peak marked with an asterisk belongs to an unidentified species. b CW X-band EPR spectra of CO-inhibited (Hox–CO) TmHydABC and TmHydA measured at 20 K and 0.1 mW microwave power. Approximately, 150 μM of each protein in 0.1 M Tris–HCl buffer pH 8, 0.15 M NaCl, 20% glycerol, were purged for 20 min with 100% CO, before measuring the spectra. The experimental spectra are overlaid with the spectral simulations (dotted magenta line) and the components are shown underneath
FTIR spectra of CO-inhibited TmHydABC and TmHydA are virtually identical and are characterized by the same IR bands at 2092 and 2085 cm−1 for the CN− ligands; 2008, 1970 and 1961 cm−1 for the terminal CO ligands and 1807 cm−1 for the bridging CO ligand. The FTIR bands of the CO ligands are in very good agreement with those recently published for native TmHydABC [30]. Both the CN− and CO IR-stretching frequencies are very similar to those reported for the Hox–CO state of other [FeFe] hydrogenases (Table S2).
The experimental EPR spectra of CO-inhibited TmHydABC and TmHydA indicate the presence of two paramagnetic species. The almost axial EPR spectra with g values [2.064, 2.008, 2.005] for TmHydABC and [2.062, 2.011, 2.006] for TmHydA are similar to those from the Hox–CO state of other [FeFe] hydrogenases (Table S3) and arise from the low spin iron centers in the mixed valence [Fe(I)Fe(II)]H, analogous to the Hox state. The oxidized sample of TmHydABC isolated from the native organism showed features at g = 2.070, 2.024, and 2.002 [26]. The EPR lines at 2.070 and 2.002 are similar to the Hox–CO EPR signature observed here for artificially maturated TmHydABC or for other [FeFe] hydrogenases. The second paramagnetic species observed in the EPR spectra of the Hox–CO state can be assigned to a reduced [2Fe–2S] cluster, as it has g values and line shapes similar to those observed for the third component in the EPR spectra of the Hox samples.
Characterization of the reduced state
Reduction of the Fe-centers in the H-cluster causes red-shifts of the FTIR peaks (with respect to Hox) of the CO and CN− ligands due to increases of electron density in anti-bonding ligand orbitals, which lengthens the CO and CN− bonds [43]. This effect is largest when reduction takes place at the [2Fe]H subcluster (20–50 cm−1); however, small red-shifts (≈10 cm−1) are also observable when the [4Fe–4S]H subcluster is reduced [8]. When reduced with sodium dithionite, FTIR spectra of TmHydABC and TmHydA are identical and showed five major IR bands at 2075, 2037, 1956, 1919, and 1887 cm−1 (Fig. 7a). All these bands are significantly red-shifted (by 20–50 cm−1) as compared to the Hox state indicating reduction of the [2Fe]H subsite. They resemble closely those for the HredH+ state identified in other [FeFe] hydrogenases (see Table S2). It is to be noted here that in the HredH+ state, the bridging CO vibration is absent in both TmHydABC and TmHydA; however, an additional peak is observed at 1956 cm−1 (Fig. 7a), which may indicate that in the HredH+ state the bridging CO becomes terminal, as was proposed for the HredH+ state in the [FeFe] hydrogenases from Chlamydomonas reinhardtii (CrHydA1) and Desulfovibrio desulfuricans (DdHydAB) [8, 44, 45]. However, in a recent study, it was suggested that the bridging CO can be retained in the HredH+ state under certain conditions [46].
FTIR and EPR spectra of the reduced TmHydABC and TmHydA. a FTIR spectra of reduced TmHydABC or TmHydA samples are shown in the upper and lower panels, respectively. Approximately 400 µM of ‘as-isolated’ protein samples were incubated with 20 mM of sodium dithionite (NaDT) at room temperature for 5 min before measuring the spectra at room temperature. The peaks corresponding to the HredH+ state are shaded in green. The minor peaks shaded in blue and red belong to the Hox–CO and Hox states. The peak marked with an asterisk belongs to an unidentified species. b CW X-band EPR spectra of reduced protein samples (TmHydABC and TmHydA) were measured at 10 K (0.01 mW microwave power) and 40 K (1 mW microwave power). EPR sample composition: 150 μM, 10 mM of NaDT, 0.1 M Tris–HCl buffer pH 8, 0.15 M NaCl, 20% glycerol
Due to the antiferromagnetic spin coupling in the [Fe(I)Fe(I)] unit, the H-cluster in the HredH+ state is EPR silent, but the reduced F-clusters give rise to a characteristic EPR spectrum consisting of multiple contributions from [4Fe–4S]+ and [2Fe–2S]+ clusters (Fig. 7b) very similar to the EPR spectrum observed for the reduced apo-enzyme (Figure S5B) and that of the reduced native enzyme [26, 38, 47]. Interestingly, the dominant contribution with g values ([2.004, 1.950, 1.920] for TmHydABC and [2.004, 1.955, 1.923] for TmHydA) is also present in the Hox and Hox–CO states (Figs. 5b, 6b) and was tentatively assigned to one of the two [2Fe–2S]+ clusters in the HydA subunit.
Spectroelectrochemical characterization of TmHydABC
Spectroelectrochemical FTIR was used previously in studies of several [FeFe] hydrogenases to investigate their equilibrium redox properties and to calculate midpoint potentials [7, 44]. Here, we apply this method to characterize the potential dependence of the observed redox states in TmHydABC. The reductive titration of TmHydABC was initiated at an open-circuit potential (OCP) of ≈ − 230 mV, vs SHE. The FTIR spectrum recorded at this potential suggested that the protein is mainly in the Hox state (Fig. 8a). As the potential of the cell was decreased, the peaks corresponding to the Hox state decreased in intensity and were replaced by the peaks that were previously observed in the dithionite reduced enzyme (Fig. 8a). However, when the potential was decreased lower than ≈−500 mV, the intensity of these peaks decreased but no new peaks appeared, indicating irreversible degradation of the H-cluster (Fig. 8a, b).
Spectroelectrochemical FTIR of TmHydABC. a Selected FTIR spectra recorded at − 233, − 433, − 493 and − 553 mV are shown. The experiment was performed with ≈1 mM TmHydABC in 200 mM phosphate buffer (pH 8) and 200 mM KCl with redox mediators at 15 °C. b Changes in FTIR absorbance with changing electrode potential, during reductive titration, at peak position 1939 cm−1 (Hox) and 1887 cm−1 (HredH+) are shown by red and green circles, respectively. The solid red and green lines represent the Nernst-fit corresponding to the model shown in c
The reductive titration curve obtained by plotting the intensities of the bands at 1939 cm−1 (Hox) and 1887 cm−1 (HredH+) vs. the applied potential could be fitted using the Nernst equation corresponding to a one-electron reduction (Fig. 8b, c). This fitting revealed that the state characterized by bands at 2075, 2037, 1956, 1919, and 1886 cm−1 is indeed one electron reduced with respect to the Hox state, and further supporting that this state is HredH+. The redox potential associated with the Hox/HredH+ transition for TmHydABC at pH 8 (Fig. 8b) was found to be − 420 ± 5 mV, which is similar to that reported for DdHydAB [44, 45].
The HredH+ state is formed by proton coupled electron transfer (PCET) of the Hox state [8]. In CrHydA1, which lacks the F-clusters, both Hred and HredH+ can be observed at neutral pH and the apparent pKa of the ADT amine is around 7.2 [8]. In contrast, for DdHydAB which harbors two F-clusters, the apparent pKa was found to be 9.0 [45]. This difference is explained by the influence of the proximal F-cluster on the redox potential of the [4Fe–4S]H subcluster. Redox anti-cooperativity (redox interaction between the F- and H-clusters that decreases the reduction potential of the H-cluster when the proximal F-cluster is reduced and vice versa), disfavors reduction of the [4Fe–4S]H subcluster, thus promoting direct PCET from the proximal F-cluster to the [2Fe]H subcluster upon reduction of the H-cluster. This mechanism increases the apparent pKa of the ADT amine moiety [45]. During the spectroelectrochemical titration of TmHydABC at pH 8, we did not observe any Hred state (Fig. 8). The Hred state was also not observed when the FTIR spectrum of dithionite reduced TmHydABC was measured at pH 10 (Figure S12). The exceptionally high apparent pKa of the ADT amine in TmHydABC could be the result of structural modifications in the H-cluster binding pocket with respect to DdHydAB and CrHydA1 that reinforce the effect of redox anti-cooperativity with the proximal F-cluster(s).
Artificial maturation with the unprotonatable [2Fe]ADT-analogue [2Fe]PDT [48, 49] confirmed that an unprotonated reduced state of TmHydABC is indeed accessible. The [2Fe]PDT-maturated TmHydABC exhibits an Hred state with very low signal intensity and broadened peaks (Figure S13). The reason for this is unclear, but may be related to some conformational freedom of the [2Fe] site in this enzyme. Furthermore, we found no evidence for the formation of a two electron reduced state (the HsredH+ state) in our spectroelectrochemistry data (Fig. 8). Formation of the HsredH+ state was proposed to be essential for CrHydA1 during H2 production [9]. For DdHydAB, an [FeFe] hydrogenase containing F-clusters, it was proposed that it can avoid the formation of the HsredH+ state, as the nearby F-clusters can accept the second electron and can tunnel it quickly to the H-cluster during catalysis [45]. A similar phenomenon is also likely to be operative in TmHydABC leading to the absence of HsredH+, although the protein is catalytically highly active. It should be noted that in a recent kinetics study on native TmHydABC, the bands assigned to HredH+ in our study were attributed to the doubly reduced state HsredH+ with some contribution from HredH+ (referred to as “Hsred” and “Hred”, respectively) [30]. We believe that as the positions of the FTIR bands of the HredH+ and HsredH+ states are very similar, correct assignment of bands to these two states is difficult without electrochemical titrations. The main conclusions of this kinetics study, however, are not affected by this misassignment [30]. The protonated reduced state did occur under proton reduction conditions and was identified as a catalytically active state [30].
Discussion
In this study, the heterotrimeric electron-bifurcating [FeFe] hydrogenase from T. maritima, TmHydABC, and its catalytic subunit TmHydA was produced by recombinant expression and artificial maturation. With artificial redox partners, semisynthetically produced TmHydABC and TmHydA showed significantly higher H2 production (550 ± 50 U mg−1 and 475 ± 60 U mg−1) and H2 oxidation activities (1300 ± 140 and 2000 ± 200 U mg−1) at 70 °C compared to the enzyme isolated from the native organism [26]. Native TmHydABC was previously found to use both reduced ferredoxin and NADH as electron donors for H2 production, thereby acting as an electron-confurcating hydrogenase [14]. Here, we show that the semisynthetically produced holo-TmHydABC could catalyze the reverse reaction, i.e., H2-dependent reduction of ferredoxin only in the presence of NAD+, demonstrating that the enzyme is also capable of the electron bifurcation reaction.
We have used FTIR and EPR spectroscopy to analyze the salient features of the H-cluster in TmHydABC and TmHydA under various conditions. Under oxidizing conditions, both TmHydABC and TmHydA showed spectral properties (positions of the FTIR peaks and EPR g values) similar to the Hox state of prototypical [FeFe] hydrogenases (non-electron bifurcating) (Fig. 5, Tables S1 and S2). Furthermore, when we treated TmHydABC and TmHydA with CO, FTIR, and EPR spectroscopic features of the Hox–CO state could be identified. This observation implies that like prototypical [FeFe] hydrogenases, the H-cluster of electron-bifurcating hydrogenases is also inhibited by CO. All these results suggest that properties of the H-cluster of TmHydABC are similar to those of the prototypical [FeFe] hydrogenases. This was expected as the amino acid sequences surrounding the H-cluster pocket are well conserved in electron-bifurcating and non-electron-bifurcating [FeFe] hydrogenases [50]. However, the previously published EPR spectroscopic analysis of native TmHydABC did not identify the characteristic EPR signal of the Hox state [26], which led to the speculation that the H-cluster of electron-bifurcating enzymes is different from prototypical enzymes [51].
Electron bifurcation refers to the process of splitting the electrons from a single electron donor to two different electron acceptors, one with higher redox potential and the other with lower redox potential than that of the electron donor [16, 17]. Electron-bifurcating enzymes usually contain at least one flavin along with several other redox centers and it is presumed that the bifurcation reaction happens at this flavin as this cofactor is capable of both one and two electron transfer reactions (flavin-based electron bifurcation, FBEB) [16, 51]. In general, the flavin centers responsible for FBEB show special redox properties [52]. These flavins display ‘crossed’ redox potentials: they undergo 2e− reduction from the flavoquinone state to the flavohydroquinone state and, then, upon 1e− oxidation, form a highly reducing flavosemiquinone state that can transfer electrons to low potential electron acceptors.
FBEB has been suggested to operate in TmHydABC and other electron-bifurcating [FeFe] hydrogenases [23]. Protein sequence analysis of TmHydABC and other electron-bifurcating [FeFe] hydrogenases indicates the presence of one FMN-binding site in HydB. Based on the sequence similarity between HydB and the NuoF (Nqo1) subunit of respiratory complex I [23], whose FMN does not engage in electron bifurcation, it seems unlikely that the FMN in HydB would be the site of electron bifurcation. Thus, the involvement of a second flavin for electron bifurcation was proposed in TmHydABC [23]. Here, we showed that TmHydABC containing 0.2 FMN per heterotrimer catalyzed the electron bifurcation reaction with a rate of 28% of that in the presence of added excess FMN. This fits with the idea that only one FMN per trimer is necessary for electron bifurcation. If two FMN sites were present then the 0.2 FMN would be split between the two sites, with 0.1 FMN per site (or 10% occupancy of each site) assuming equal affinity for both sites. By the law of probabilities then, only 1% of HydABC would have both sites occupied. If both FMN sites were essential for the catalytic mechanism (one for NADH oxidation and one for electron bifurcation), then, in this scenario, the addition of excess FMN should fill up all empty FMN sites and increase the activity accordingly by about 100-fold. Therefore, the modest fourfold increase in activity, contradicts the proposition of a second flavin being the bifurcation center in TmHydABC.
More recently, an alternative model of the bifurcation mechanism in [FeFe] hydrogenases has been postulated by Peters et al. where the H-cluster was hypothesized to be the electron bifurcation site [51]. The rationale behind this hypothesis was that the H-cluster fulfills one of the key requirements of being an electron-bifurcation center, as it is capable of 2e− redox reactions and can exist in different oxidation states. Like electron-bifurcating flavins, the intermediate redox state of the H-cluster should be strongly reducing in order for it to be an electron bifurcation center [52]. Our spectroelectrochemical analysis of TmHydABC shows that the H-cluster forms a stable 1e− reduced state (HredH+). The 2e− reduced state (HsredH+) was not observed under our experimental conditions; possibly due to a very low redox potential of the HredH+ ⇆ HsredH+ transition imposed by redox anti-cooperativity between the H-cluster and the reduced F-clusters [45]. This observation implies that the H-cluster does not show ‘crossed-over’ redox behavior similar to electron-bifurcating flavins. Although according to Zhang et al. redox centers with uncrossed redox potentials can also theoretically act as bifurcating centers under certain conditions [55].
A possible mechanism would be that H2 oxidation at the H-cluster produced a highly reducing HsredH+ species, which quickly transfers an electron to a nearby iron–sulfur cluster with a very negative redox potential. This would then be followed by downhill electron transfer to the ferredoxin-binding site. The HredH+ state would then transfer an electron to a different iron–sulfur cluster with a more positive redox potential. We think that this is rather unlikely to be the case in TmHydABC, due to the high sequence similarity between TmHydA and the structurally characterized [FeFe] hydrogenase from C. pasteurianum (CpI or CpHydA) [4, 54]. CpHydA contains four accessory iron–sulfur clusters, but only one of them is within electron transfer distance of the H-cluster. Thus, a second proximal iron–sulfur cluster in TmHydA seems unlikely.
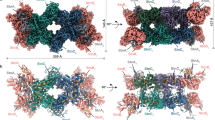
We hypothesize, instead, that a complicated arrangement of iron–sulfur clusters, and interactions between them may facilitate a novel elegant electron-bifurcating mechanism. The arrangement of cofactors and subunits in TmHydABC remains unknown. However, the strong homology between TmHydA, TmHydB, and TmHydC, and the complex I subunits TtNqo1, TtNqo2, and TtNqo3 from Thermus thermophilus [53], may indicate a similar arrangement of subunits and cofactors (Fig. 9). In this arrangement, HydC would be located on one side of HydB, positioning the [2Fe–2S] cluster of HydC close to the FMN cofactor of HydB. Meanwhile, HydA would be located on the opposite side of HydB with the surface exposed [2Fe–2S] cluster of HydA in electrical contact with the surface exposed [4Fe–4S] cluster of HydB. This arrangement would not be compatible with the previously proposed Fd-binding site being HydC. Instead, Fd would interact with the His-ligated [4Fe–4S] cluster of HydA, as has been proposed for the CpHydA [56]. With the H-cluster in one direction, the FMN in another direction and the Fd-binding site in a third direction, this arrangement temptingly implicates the trinity of iron–sulfur clusters in HydA as a potential bifurcation site. How, such an arrangement could operate to regulate electron transfer from the H-cluster in one direction or another is unclear, but we speculate that the His-ligated [4Fe–4S] cluster in HydA could play an important role. Further investigations are underway to investigate this possibility. Crucially, our recombinant method for producing TmHydABC represents the perfect system to perform such in-depth mechanistic studies of the electron-bifurcating mechanism, since it provides an efficient way to produce very high yields of pure protein, as well as making it easy to produce site directed mutations to directly test these ideas.
Proposed arrangement of subunits and cofactors in TmHydABC. The TmHydABC subunits are homologous to the Nqo1, Nqo2, and Nqo3 subunits of the structurally characterized complex I from Thermus thermophilus. Based on this homology, the arrangements of the conserved cofactors can be predicted. The figure shows the protein subunits Nqo1 (HydB, green), Nqo2 (HydC, blue), and Nqo3 (HydA, pink) in the cartoon representation (PDB: 4HEA [53],), with the cofactors from Nqo1 and Nqo2 from complex I, and the cofactors from the [FeFe] hydrogenase CpHydA (PDB: 4XDC [54]), overlaid. CpHydA was aligned to Nqo3 in Pymol giving almost perfect alignment of the homologous clusters. HydA contains an additional [2Fe–2S] cluster, for which CpHydA does not contain a homologous cluster, and HydB contains an additional two [4Fe–4S] clusters and one [2Fe–2S] cluster, for which complex I does not contain homologous clusters
Conclusion
In this study, we have developed a method of producing the heterotrimeric electron-bifurcating [FeFe] hydrogenase from T. maritima using recombinant expression and artificial maturation. The time efficiency of the recombinant expression method prevented protein damage and led to high catalytic activity for both TmHydABC and TmHydA, outperforming enzymes isolated from the native organism. Our preparation was competent in the electron bifurcation reaction, even in the absence of added FMN. Using FTIR and EPR spectroscopy the three typical states present in all active [FeFe] hydrogenases, i.e., Hox, HredH+, and Hox–CO could be identified in both TmHydABC and TmHydA. The unprotonated singly reduced state Hred as well as the doubly reduced state HsredH+ (both with a reduced [4Fe–4S]-subcluster) were not observed under any condition. This is taken as evidence for a strong electronic coupling between the H-cluster and the F-clusters in the enzyme disfavoring reduction of the cubane subcluster. Our results do not agree with the FMN or the H-cluster as being the site of electron bifurcation. Instead, we hypothesize that the iron–sulfur clusters in the HydA subunit could serve this function. The efficient method presented here for obtaining high quantities of high quality TmHydABC should pave the road for more protein intensive experiments such as X-ray crystallography, spectroelectrochemistry, and nuclear resonance vibrational spectroscopy, to test our hypothesis, and to help understand the enigmatic mechanism of electron bifurcation in this fascinating [FeFe] hydrogenase.
Abbreviations
- NADH:
-
Dihydronicotinamide adenine dinucleotide
- NAD+ :
-
Nicotinamide adenine dinucleotide
- Fdox :
-
Oxidized ferredoxin
- Fdred :
-
Reduced ferredoxin
- ADT:
-
2-azapropane 1,3-dithiolate
- FMN:
-
Flavin mononucleotide
- FTIR:
-
Fourier transform infrared
- EPR:
-
Electron paramagnetic resonance
- FAD:
-
Flavin adenine dinucleotide
- CW:
-
Continuous wave
- OCP:
-
Open-circuit potential
- PCET:
-
Proton-coupled electron transfer
- FBEB:
-
Flavin-based electron bifurcation
References
Lubitz W, Ogata H, Rüdiger O, Reijerse E (2014) Chem Rev 114:4081–4148
Peters JW, Schut GJ, Boyd ES, Mulder DW, Shepard EM, Broderick JB, King PW, Adams MW (2015) Biochim Biophys Acta 1853:1350–1369
Glick BR, Martin WG, Martin SM (1980) Can J Microbiol 26:1214–1223
Peters JW, Lanzilotta WN, Lemon BJ, Seefeldt LC (1998) Science 282:1853–1858
Nicolet Y, Piras C, Legrand P, Hatchikian CE, Fontecilla-Camps JC (1999) Structure 7:13–23
Winkler M, Esselborn J, Happe T (2013) Biochim Biophys Acta 1827:974–985
Silakov A, Kamp C, Reijerse E, Happe T, Lubitz W (2009) Biochemistry 48:7780–7786
Sommer C, Adamska-Venkatesh A, Pawlak K, Birrell JA, Rüdiger O, Reijerse EJ, Lubitz W (2017) J Am Chem Soc 139:1440–1443
Adamska A, Silakov A, Lambertz C, Rüdiger O, Happe T, Reijerse E, Lubitz W (2012) Angew Chem Int Ed Engl 51:11458–11462
Mulder DW, Guo Y, Ratzloff MW, King PW (2017) J Am Chem Soc 139:83–86
Reijerse EJ, Pham CC, Pelmenschikov V, Gilbert-Wilson R, Adamska-Venkatesh A, Siebel JF, Gee LB, Yoda Y, Tamasaku K, Lubitz W, Rauchfuss TB, Cramer SP (2017) J Am Chem Soc 139:4306–4309
Greening C, Biswas A, Carere CR, Jackson CJ, Taylor MC, Stott MB, Cook GM, Morales SE (2016) ISME J 10:761–777
Sondergaard D, Pedersen CN, Greening C (2016) Sci Rep 6:34212
Schut GJ, Adams MW (2009) J Bacteriol 191:4451–4457
Huang H, Wang S, Moll J, Thauer RK (2012) J Bacteriol 194:3689–3699
Buckel W, Thauer RK (2018) Chem Rev 118:3862–3886
Peters JW, Miller AF, Jones AK, King PW, Adams MW (2016) Curr Opin Chem Biol 31:146–152
Mitchell P (1975) FEBS Lett 59:137–139
Li F, Hinderberger J, Seedorf H, Zhang J, Buckel W, Thauer RK (2008) J Bacteriol 190:843–850
Muller V, Chowdhury NP, Basen M (2018) Annu Rev Microbiol 72:331–353
Zheng YN, Kahnt J, Kwon IH, Mackie RI, Thauer RK (2014) J Bacteriol 196:3840–3852
Buckel W, Thauer RK (2018) Front Microbiol 9:401
Buckel W, Thauer RK (2013) Biochim Biophys Acta 1827:94–113
Nakamura N, Lin HC, McSweeney CS, Mackie RI, Gaskins HR (2010) Annu Rev Food Sci Technol 1:363–395
Wolf PG, Biswas A, Morales SE, Greening C, Gaskins HR (2016) Gut Microbes 7:235–245
Verhagen MF, O’Rourke T, Adams MW (1999) Biochim Biophys Acta 1412:212–229
Schuchmann K, Muller V (2012) J Biol Chem 287:31165–31171
Wang S, Huang H, Kahnt J, Thauer RK (2013) J Bacteriol 195:1267–1275
Kpebe A, Benvenuti M, Guendon C, Rebai A, Fernandez V, Le Laz S, Etienne E, Guigliarelli B, Garcia-Molina G, de Lacey AL, Baffert C, Brugna M (2018) Biochim Biophys Acta Bioenerg 1859:1302–1312
Greene BL, Schut GJ, Adams MWW, Dyer RB (2017) ACS Catal 7:2145–2150
Pan G, Menon AL, Adams MW (2003) J Biol Inorg Chem 8:469–474
Verhagen MF, O’Rourke TW, Menon AL, Adams MW (2001) Biochim Biophys Acta 1505:209–219
Birrell JA, Laurich C, Reijerse EJ, Ogata H, Lubitz W (2016) Biochemistry 55:4344–4355
Esselborn J, Lambertz C, Adamska-Venkatesh A, Simmons T, Berggren G, Noth J, Siebel J, Hemschemeier A, Artero V, Reijerse E, Fontecave M, Lubitz W, Happe T (2013) Nat Chem Biol 9:607–609
Birrell JA, Rüdiger O, Reijerse EJ, Lubitz W (2017) Joule 1:61–76
Moss D, Nabedryk E, Breton J, Mantele W (1990) Eur J Biochem 187:565–572
Stoll S, Schweiger A (2006) J Magn Reson 178:42–55
Juszczak A, Aono S, Adams MW (1991) J Biol Chem 266:13834–13841
Kertess L, Wittkamp F, Sommer C, Esselborn J, Rüdiger O, Reijerse EJ, Hofmann E, Lubitz W, Winkler M, Happe T, Apfel UP (2017) Dalton Trans 46:16947–16958
Wang S, Huang H, Kahnt J, Mueller AP, Kopke M, Thauer RK (2013) J Bacteriol 195:4373–4386
Lemon BJ, Peters JW (1999) Biochemistry 38:12969–12973
Pierik AJ, Hulstein M, Hagen WR, Albracht SP (1998) Eur J Biochem 258:572–578
Lubitz W, Reijerse E, van Gastel M (2007) Chem Rev 107:4331–4365
Roseboom W, De Lacey AL, Fernandez VM, Hatchikian EC, Albracht SP (2006) J Biol Inorg Chem 11:102–118
Rodríguez-Maciá P, Pawlak K, Rüdiger O, Reijerse EJ, Lubitz W, Birrell JA (2017) J Am Chem Soc 139:15122–15134
Ratzloff MW, Artz JH, Mulder DW, Collins RT, Furtak TE, King PW (2018) J Am Chem Soc 140:7623–7628
Smith ET, Adams MW (1994) Biochim Biophys Acta 1206:105–112
Siebel JF, Adamska-Venkatesh A, Weber K, Rumpel S, Reijerse E, Lubitz W (2015) Biochemistry 54:1474–1483
Adamska-Venkatesh A, Krawietz D, Siebel J, Weber K, Happe T, Reijerse E, Lubitz W (2014) J Am Chem Soc 136:11339–11346
Poudel S, Tokmina-Lukaszewska M, Colman DR, Refai M, Schut GJ, King PW, Maness PC, Adams MW, Peters JW, Bothner B, Boyd ES (2016) Biochim Biophys Acta 1860:1910–1921
Peters JW, Beratan DN, Schut GJ, Adams MWW (2018) Chem Commun 54:4091–4099
Nitschke W, Russell MJ (2012) BioEssays 34:106–109
Baradaran R, Berrisford JM, Minhas GS, Sazanov LA (2013) Nature 494:443–448
Esselborn J, Muraki N, Klein K, Engelbrecht V, Metzler-Nolte N, Apfel UP, Hofmann E, Kurisu G, Happe T (2016) Chem Sci 7:959–968
Zhang P, Yuly JL, Lubner CE, Mulder DW, King PW, Peters JW, Beratan DN (2017) Acc Chem Res 50:2410–2417
Artz JH, Mulder DW, Ratzloff MW, Lubner CE, Zadvornyy OA, LeVan AX, Williams SG, Adams MWW, Jones AK, King PW, Peters JW (2017) J Am Chem Soc 139:9544–9550
Acknowledgements
Open access funding provided by Max Planck Society. We thank Yvonne Brandenburger for helping with sample preparation and biochemical characterization and Tabea Mußfeld for synthesis of the complexes. We also thank Norbert Dickmann (MALDI TOF–MS), Dr. Philip Schulze and Sylvia Ruthe, MPI-Kohlenforschung (gas chromatography), and Ingeborg Heise (synthesis) for their assistance. The work was supported by the Max Planck Society and in part by JSPS KAKENHI Grant number 16K21748 (H. O.). J. B. acknowledges the Deutsche Forschungsgemeinschaft Priority Programme “Iron–Sulfur for Life: Cooperative Function of Iron–Sulfur Centers in Assembly, Biosynthesis, Catalysis and Disease” (SPP 1927) Project BI 2198/1-1.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chongdar, N., Pawlak, K., Rüdiger, O. et al. Spectroscopic and biochemical insight into an electron-bifurcating [FeFe] hydrogenase. J Biol Inorg Chem 25, 135–149 (2020). https://doi.org/10.1007/s00775-019-01747-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00775-019-01747-1