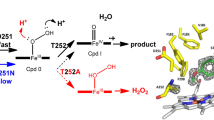
Abstract
The Thr252 residue plays a vital role in the catalytic cycle of cytochrome P450cam during the formation of the active species (Compound I) from its precursor (Compound 0). We investigate the effect of replacing Thr252 by methoxythreonine (MeO-Thr) on this protonation reaction (coupling) and on the competing formation of the ferric resting state and H2O2 (uncoupling) by combined quantum mechanical/molecular mechanical (QM/MM) methods. For each reaction, two possible mechanisms are studied, and for each of these the residues Asp251 and Glu366 are considered as proton sources. The computed QM/MM barriers indicate that uncoupling is unfavorable in the case of the Thr252MeO-Thr mutant, whereas there are two energetically feasible proton transfer pathways for coupling. The corresponding rate-limiting barriers for the formation of Compound I are higher in the mutant than in the wild-type enzyme. These findings are consistent with the experimental observations that the Thr252MeO-Thr mutant forms the alcohol product exclusively (via Compound I), but at lower reaction rates compared with the wild-type enzyme.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cytochrome P450 is one of the most versatile enzymes in nature [1, 2]. It uses dioxygen to catalyze a great variety of stereospecific and regioselective processes of oxygen insertion into organic compounds [3–8]. These processes are of vital importance in biosystems, where the enzyme participates in detoxification and in biosyntheses [1]. Since the activation of inert C–H bonds is one of the holy grails of chemistry [9], the facility to carry out this process makes the P450 enzyme superfamily a model for creative mimetic chemistry [10] designed to generate novel catalysts that can perform C–H activation.
The bacterial enzyme P450cam (CYP101) is the workhorse of P450 research which has generated much insight into the role of the protein in regulating the activity of the enzyme and the effects of site-directed mutations [4, 11–14]. Its active site contains a heme unit that consists of an iron protoporphyrin IX complex with Cys as the proximal axial ligand. The active catalytic species, with an Fe(IV)–oxo moiety, is commonly denoted as Compound I. It has been observed in a related chloroperoxidase, but is still elusive for P450 enzymes, where it has only been identified by transient spectroscopy [8].
Site-directed mutagenesis studies [15–18] in combination with X-ray structural analyses [19, 20] indicate that the conserved P450 residue Thr252 at the active site plays a crucial role in the catalysis, in particular during the formation of Compound I [21, 22]. Hence, it was no surprise that Thr252 became an early target for mutagenesis [23, 24]. Several mutants such as Thr252Ala and Thr252Gly show an uncoupling of O2 consumption from d-camphor hydroxylation, most of the O2 consumed being converted to H2O2 without cleaving the O–O bond (Scheme 1), whereas Thr252Ser retains significant coupling of O2 consumption with d-camphor hydroxylation [15]. When Thr252 is replaced by methoxythreonine (MeO-Thr), the resulting Thr252MeO-Thr mutant gives 100% formation of 5-exo-hydroxycamphor (no uncoupling), but the rate of reaction is slowed down to one third compared with that for the wild-type enzyme [25]. None of the other Thr252X mutants studied preserves the coupling activity to a similar extent [25]. It is generally assumed that site-directed Thr252X mutagenesis will disrupt the proton relay that converts Compound 0 to Compound I in P450 enzymes [26, 27].
Several theoretical studies have addressed the protonation reactions that generate Compound I [28–37]. The commonly formulated mechanism is protonation of Compound 0 at the distal oxygen atom followed by O–O bond cleavage. Recent density functional theory calculations on a large gas-phase active-site model (96 atoms) indeed gave a stable protonated Compound 0 species with significant barriers for the conversion toward both Compound 0 and Compound I [32]. However, subsequent quantum mechanical/molecular mechanical (QM/MM) calculations showed that such an intermediate is quite instable in the enzyme (more than 20 kcal/mol above Compound 0) and mechanistically irrelevant (barriers of only 3–4 kcal/mol for the decay to Compound 0 and Compound I) [37]. An alternative mechanism was proposed that involves an initial O–O bond cleavage followed by a proton transfer to the OH species formed (via a hydrogen-bonding network in the Asp251 channel), with a concomitant electron transfer from the heme (yielding Compound I and water) [37]. A similar mechanistic scenario was considered in heme oxygenase and chloroperoxidase [38, 39]. The latest QM/MM work [40] on this topic investigated both the coupling and the uncoupling reactions of Compound 0 in the wild-type P450cam enzyme and in four Thr252X mutants (X = Ser, Val, Ala, Gly). It was found that the formation of Compound I (coupling) always proceeds through the two-step mechanism with initial O–O bond cleavage [40]. By contrast, the uncoupling reaction is always concerted. Its barrier is always higher than that of the coupling reaction if the Asp251 channel contains only residue 252, the crystallographic water molecule Wat901, and protonated Asp251. Molecular dynamics (MD) simulations indicate, however, that an additional water molecule is stable in the Asp251 channel for X = Val, Ala, and Gly, which leads to much smaller barriers for uncoupling owing to a much more favorable hydrogen-bonding network. Including this extra water molecule in the QM region makes uncoupling competitive with coupling in the case of X = Val and renders it more facile for X = Ala and Gly [40], in qualitative agreement with experiment [15].
Here, we extend our previous QM/MM work by considering the effect of the Thr252MeO-Thr mutation. We address both the coupling and the uncoupling reactions and attempt to answer the question whether the Thr252MeO-Thr mutation will indeed disrupt the proton relay channel that is commonly viewed as being an essential prerequisite for the conversion of Compound 0 to Compound I.
Computational methods and proposed mechanisms
The initial structure was taken from the MD trajectory of the native enzyme studied earlier [37]. Thr was mutated into MeO-Thr by manually replacing the OH group in the Thr252 residue by OCH3. The same solvation and protonation schemes were applied as in previous studies [41–43]. Glu366 and Asp251 were considered as possible proton sources [17, 31, 44], and the corresponding two protonation schemes were adopted as in the standard setup used previously [41–43] (i.e., protonated Glu366 and deprotonated Asp251 in the Glu366 channel, and deprotonated Glu366 and protonated Asp251 in the Asp251 channel). Both setups consisted of 24,988 atoms in total, including 5,891 TIP3P water molecules [45]. The solvated systems were relaxed by performing classical energy minimizations and MD simulations at the MM level using the CHARMM22 force field [46] as implemented in the CHARMM program [47]. The heme units with the Cys357 and OOH ligands as well as the outer 8 Å of the solvent layer were kept fixed during these initial runs.
The QM/MM method chosen was analogous to that used in our previous studies [41–43]. Here, we briefly mention some aspects relevant to the present work. Minimized snapshots from the MD trajectories were taken as initial structures for QM/MM optimizations. In the QM/MM calculations, the QM part was treated by unrestricted hybrid density functional theory (UB3LYP) [48] with the LACVP [49] small-core effective core potential basis set on iron and 6-31G [50] on the remaining atoms (B1) for geometry optimizations, while the MM part was described by the CHARMM22 force field. Single-point calculations were carried out with the TZVP [51, 52] basis set (B2).
An electronic embedding scheme [53] was adopted in the QM/MM calculations, i.e., interactions with MM charges were incorporated into the one-electron Hamiltonian of the QM calculation. No cutoffs were introduced for the nonbonding MM and QM/MM interactions. Hydrogen link atoms with the charge shift model were employed to treat the QM/MM boundary. The TURBOMOLE program [54] was used for the QM treatment in the QM/MM calculations as well as in pure QM calculations. The CHARMM22 force field was run through the DL_POLY [55] program to handle the MM part of the systems. The QM/MM calculations were performed with the ChemShell package [56], which integrates the TURBOMOLE and DL_POLY programs and performs geometry optimization with the HDLC optimizer [57].
Possible proton transfer pathways
Scheme 1 shows the four proposed mechanisms that were investigated for both protonation channels (Glu366 and Asp251).
In mechanism I, initially the O–O bond is cleaved to generate an OH radical and one-electron-reduced Compound I. Subsequently, a proton is transferred to the OH radical with a concomitant electron transfer from the heme that yields Compound I and water [37].
In mechanism II, a proton is transferred to the distal oxygen atoms of the hydroperoxo group to form protonated Compound 0 (containing FeOOH2), followed by heterolytic O–O bond cleavage that generates Compound I and water.
Mechanisms I and II both give Compound I and correspond to the coupling reaction (Scheme 1a).
In mechanism III, initially the Fe–O bond is cleaved to generate an OOH radical, followed by a proton transfer to the OOH group that yields the ferric resting state and hydrogen peroxide.
In mechanism IV, a proton is transferred to the proximal oxygen atom of the hydroperoxo group to form an FeH2O2 moiety, followed by heterolytic cleavage of the O–Fe bond generating the ferric resting state and hydrogen peroxide.
Mechanisms III and IV both yield the ferric resting state and hydrogen peroxide (uncoupling reaction, Scheme 1b).
QM region
In the QM/MM calculations, we employed QM regions analogous to those adopted for the wild-type enzyme in our previous work [37] (Fig. 1), except that the Thr252 residue was replaced by MeO-Thr. In both channels (Asp251 and Glu366), the QM region included: iron porphine (without heme side chains), the sulfur atom of Cys357, the axial OOH moiety, and MeO-Thr (represented by CH3OCH2CH3). In addition, the QM region also contained Wat901 and Asp251 (represented by CH3COOH) in the case of the Asp251 channel, and Wat523, Wat566, Wat687, Wat902, and Glu366 (represented by CH3COOH) in the case of the Glu366 channel (Fig. 1). Hence, the water molecules that may be involved in the proton transfer are part of the QM region for each channel.
Compound 0 can exist in a doublet, quartet or sextet state. It has a doublet ground state both in the wild-type P450cam enzyme and in the Thr252X mutants. According to the QM/MM calculations, the lowest quartet and sextet states lie 8.3 and 9.0 kcal/mol above the doublet ground state of the Thr252MeO-Thr mutant, respectively. Therefore, we only studied the reactions in the doublet state of the mutant, as was done previously in the case of the wild-type enzyme [37].
Results
Figures 2, 3, 4, 5, 6, 7, and 8 show the optimized QM/MM geometries of the QM regions for all relevant minima and transition states (Thr252MeO-Thr mutant, mechanisms I–IV, Glu366 and Asp251 channels). The computed relative QM/MM energies of the stationary points are summarized for basis sets B1/B2 in Table 1 (coupling reaction, mechanisms I and II) and Table 2 (uncoupling reaction, mechanisms III and IV). The single-point energies obtained with the larger TZVP basis (B2) at the corresponding optimized QM/MM geometries (B1) are generally quite similar to those obtained with the smaller basis (B1), although they are consistently slightly higher relative to Compound 0, typically by 1–3 kcal/mol. A similar behavior was also observed for the wild-type enzyme [37]. In the following discussion, we shall only quote B1 results for the sake of consistency (energies, geometries, etc.). Formation of the correct intermediates and products was verified by analysis of the spin densities and Mulliken charges. These data and selected geometrical parameters are documented in the electronic supplementary material.
Mechanism I: homolytic O–O bond cleavage followed by coupled proton–electron transfer
Glu366 channel
The first step passes over a barrier of 18.1 kcal/mol and leads to an intermediate (IC1), in which the OH moiety forms two hydrogen bonds with MeO-Thr and with Fe=O (Fig. 2). During this step, the Fe–O bond shortens to 1.67 Å in TS1 and then remains at 1.68 Å in IC1. These structural features are similar to those reported for the wild-type enzyme [37]. The spin density and partial charge of the OH group in the first intermediate (IC1) are −0.93 and −0.04, indicating that IC1 contains an OH radical and one-electron-reduced Compound I. This suggests that the O–O bond cleavage is homolytic: the Fe=O moiety carries two unpaired electrons, and the third unpaired electron is mainly located on the OH moiety. IC1 is stabilized by hydrogen-bonding interactions of OH with FeO and MeO-Thr252, and therefore lies only 10.3 kcal/mol above the reactant.
The second step is a hydrogen transfer from the MeO-Thr group to the OH moiety which yields Compound I and water. The corresponding transition state (TS2) lies 18.5 kcal/mol above Compound 0, and the intermediate complex of CH2O-Thr radical with Compound I (IC2) is quite stable, with an energy of 1.1 kcal/mol relative to Compound 0. The OH moiety is obviously reactive enough to abstract a proton from the methoxy group, and the resulting intermediate (IC2) is stabilized by Wat902 and the water molecule formed via two hydrogen bonds.
In the last step, a proton is transported from Glu366 to MeO-Thr in a concerted process via three bridging water molecules. Simultaneously, an electron is transferred from the heme to the methylene group to regenerate the MeO-Thr and form a π cation radical at the heme. The transition state (TS3) and the product (Compound I) lie 17.2 and 8.0 kcal/mol above Compound 0, respectively. The hydrogen-bonding network between Glu366 and MeO-Thr is reoriented after the proton transfer. Overall, the rate-limiting step is the hydrogen abstraction from the methoxy group with a barrier of 18.5 kcal/mol (TS2).
Asp251 channel
In this channel, the barrier of O–O bond cleavage is 18.6 kcal/mol (TS1 in Fig. 3), similar to the corresponding barrier in the Glu366 channel (18.1 kcal/mol). The intermediate (IC1, OH moiety and one-electron-reduced Compound I) is rather high in energy (14.4 kcal/mol). For the conversion of IC1 to Compound I, a proton needs to be transported from the Asp251 carboxyl group via Wat901 and MeO-Thr to OH, with a concomitant electron transfer from the heme. The spin density and partial charge of the OH group in IC1 are −0.79 and −0.12, indicating that OH will not behave as a “perfect” radical in IC1 owing to the strong hydrogen-bonding interactions with the methoxy group (2.10 Å) and the FeO unit (1.88 Å). In contrast to the wild-type enzyme [37], the subsequent proton delivery proceeds in two steps. As in the Glu366 channel, a hydrogen atom is first transferred from the methoxy group of MeO-Thr (TS2 at 22.6 kcal/mol, i.e., 8.2 kcal/mol above IC1). The intermediate formed (IC2 at 11.4 kcal/mol) then receives a proton through the Asp251 channel and an electron from the heme in a simultaneous process (TS3 at 23.0 kcal/mol). After releasing its proton, the side chain of Asp251 rotates back into a salt bridge with Arg186, as shown in Fig. 3.
Comparison
In each channel, the three transition states lie at similar energies relative to Compound 0. The highest point in the reaction profile is TS2 (TS3) in the Glu366 (Asp251) channel at 18.5 (23.0) kcal/mol (see Table 1), i.e., about 4–8 kcal/mol higher than in the wild-type enzyme [37]. The conversion of Compound 0 to Compound I via mechanism I should thus be much slower in the Thr252MeO-Thr mutant compared with the wild-type enzyme.
Additional snapshot
To ensure that the snapshot used in this study is representative for the system, reaction mechanism I in the Asp251 channel was also studied in an analogous manner with a different snapshot which was drawn after 1,500 ps of MD simulation. The computed relative energies of all stationary points (Table S23) agree with those from the first snapshot (Table 1) to within 1 kcal/mol. The highest point in the reaction profile (TS3) is at 23.2 kcal/mol, very close to the value of 23.0 kcal/mol from the first snapshot (see earlier). The results from both snapshots are thus entirely consistent with each other.
Mechanism II: proton-assisted heterolytic O–O bond cleavage
Glu366 channel
The energy barrier for direct hydrogen atom transfer from MeO-Thr to FeOOH is 20.8 kcal/mol, and the resulting intermediate (IC1, Fig. 4) lies 0.8 kcal/mol above Compound 0. The unpaired electron is mainly located on the iron atom (iron spin density of 1.37). In contrast to the reaction in the wild-type enzyme, IC1 is not a protonated Compound 0 species, since the O–O bond is cleaved in the first step [37]. However, mechanism II differs from mechanism I, since the hydrogen transfer is part of the first step. In the second step, the concomitant transport of one proton (from Glu366) and one electron (from the heme) leads to formation of Compound I. The relative energies of TS2 and Compound I are 20.0 and 8.0 kcal/mol, respectively.
Asp251 channel
In the Asp251 channel, we chose several different reaction coordinates to convert Compound 0 to protonated Compound 0 by proton transfer from Asp251 to the distal oxygen atom of the hydroperoxo group. However, all energy scans led to continuously increasing energy profiles, and we were unable to locate protonated Compound 0. Similar problems have also been reported in previous QM/MM calculations for the wild-type enzyme [37].
Mechanism III: homolytic O1–Fe bond cleavage followed by coupled proton–electron transfer
Glu366 channel
The optimized geometries are presented in Fig. 5. The barrier (TS1) for homolytic breaking of the O1–Fe bond is 30.3 kcal/mol, and the intermediate (IC1) consisting of iron-bound heme and the OOH radical lies 17.7 kcal/mol above Compound 0. The subsequent hydrogen transfer from MeO-Thr to OOH is very difficult (TS2 at 42.4 kcal/mol, thus 24.7 kcal/mol above IC1), and the second intermediate (IC2) with iron-bound heme and the CH2O-Thr radical is a shallow minimum (IC2 at 40.6 kcal/mol). The barrier for final proton transfer from Glu366 to CH2O-Thr with concomitant electron transfer from the heme is prohibitively high (TS3 at 60.7 kcal/mol). The overall reaction is endothermic by 28.4 kcal/mol.
Asp251 channel
Figure 6 shows the optimized geometries. In general, the barriers are quite similar to those in the Glu366 channel. The barrier (TS1) for homolytic cleavage of the O1–Fe bond is 28.1 kcal/mol. In the resulting intermediate (IC1 at 25.7 kcal/mol), the spin densities of OOH (−0.97) and iron (1.98) indicate that iron has two unpaired electrons and that OOH is present as a radical. The following hydrogen transfer from MeO-Thr to OOH is again difficult (TS2 at 47.6 kcal/mol, hence 21.9 kcal/mol above IC1) and leads to a very shallow intermediate (IC2 at 47.4 kcal/mol) with a CH2O-Thr radical (spin density of −0.92). The final proton transfer from Asp251 to CH2O-Thr requires much activation (TS3 at 58.4 kcal/mol, i.e., 11.0 kcal/mol above IC2). At the end of the reaction, Asp251 rotates to rebuild the salt bridge with Arg186, as also found in mechanism I. The overall reaction is endothermic by 28.8 kcal/mol.
Mechanism IV: proton-assisted heterolytic O–Fe bond cleavage
Glu366 channel
As can be seen from Fig. 7, the first step involves O–Fe bond cleavage combined with a hydrogen transfer from MeO-Thr to the proximal oxygen atom. The corresponding barrier is high (TS1 at 40.3 kcal/mol), and the shallow intermediate (IC1 at 38.0 kcal/mol) contains essentially neutral hydrogen peroxide with almost zero spin density and an O1–O2 distance of 1.51 Å; the Fe–O1 distance increases from 1.85 Å (Compound 0) to 3.75 Å (IC1). The subsequent proton transfer from Glu366 to CH2O-Thr again needs much activation (TS2 at 58.3 kcal/mol, thus 20.3 kcal/mol above IC1). The product (ferric resting state and hydrogen peroxide) lies 25.2 kcal/mol above Compound 0.
Asp251 channel
Figure 8 presents the optimized geometries. The O–Fe bond cleavage with formation of hydrogen peroxide again occurs in the first step, which has a very high barrier (TS1 at 52.9 kcal/mol). The intermediate (IC1 at 47.6 kcal/mol) contains hydrogen peroxide and a CH2O-Thr radical (spin density of −0.90). The transition state for proton transfer from Asp251 to CH2O-Thr (TS2 at 54.9 kcal/mol) lies 7.3 kcal/mol above IC1. The overall reaction is endothermic by 26.4 kcal/mol.
Discussion and conclusions
In this work, the coupling and uncoupling reactions in the Thr252MeO-Thr mutant of cytochrome P450cam were investigated for two proton delivery channels (Glu366 and Asp251) and four possible mechanisms by means of QM/MM calculations. It is obvious from the QM/MM results that the uncoupling reaction (formation of the ferric resting state and hydrogen peroxide) is strongly disfavored. Regardless of mechanistic details, it suffers from high endothermicities of 25–30 kcal/mol and extremely high overall activation energies of 55–61 kcal/mol. We note in this context that our previous QM/MM study [37] of the uncoupling reaction in the wild-type enzyme yielded a barrier of 27 kcal/mol in the Asp251 channel, with the required proton being provided via the Asp251–Wat901–Thr252 network (mechanism similar to mechanism IV). Such proton delivery is expected to be more facile in the wild-type enzyme than the corresponding process in the Thr252MeO-Thr mutant since the hydroxyl group in Thr is a much better proton donor than the methyl group in MeO-Thr, and it is thus not surprising that the uncoupling reaction requires more activation in the Thr252MeO-Thr mutant.
In our recent QM/MM study [40], we discovered that the barrier for uncoupling is dramatically reduced in the Thr252X mutants (X = Val, Ala, Gly) when an extra water molecule enters the Asp251 channel and becomes part of a well-connected hydrogen-bonding network that provides a good proton delivery pathway. In these mutants, the extra water molecule remains present in 2-ns MD simulations, whereas it escapes from the channel for X = Ser and X = Thr. Also in the Thr252MeO-Thr mutant, the stability of an additional water molecule was tested by means of 2-ns MD simulations. It was observed that the additional water molecule escapes from the active site in both channels in the course of the MD simulations (Figs. S2, S3). This is in agreement with our previous findings for the wild-type enzyme, since MeO-Thr is sterically more demanding than Thr.
According to the present QM/MM results for the Thr252MeO-Thr mutant, the coupling reaction (mechanisms I and II) is endothermic by about 8 kcal/mol and requires an overall activation of 18–23 kcal/mol, depending on the channel and mechanism. It thus seems feasible and is clearly preferred over the uncoupling reaction. For both the coupling reaction and the uncoupling reaction, the highest point on the computed QM/MM energy profiles corresponds to hydrogen abstraction by OH and OOH species that are present in the intermediates formed. It is well known, e.g., from QM studies on small model systems [58], that such reactions are intrinsically more facile and more exothermic with OH than with OOH. For example, at the UB3LYP/6-31+G* level, the barrier (reaction energy) for hydrogen abstraction from ethyl methyl ether is 2.1 (−15.0) kcal/mol for OH and 12.6 (10.0) kcal/mol for OOH (Fig. S1). These intrinsic preferences are reflected in the QM/MM energies (Tables 1, 2).
Experimentally, the Thr252MeO-Thr mutant gives 100% coupling reaction and no uncoupling reaction [25], consistent with our QM/MM results. Furthermore, the observed rate constant for the formation of 5-exo-hydroxycamphor is one third of that of the wild-type enzyme [25]. This is in qualitative agreement with the QM/MM finding that the rate-limiting barriers for the coupling reaction are higher in the Thr252MeO-Thr mutant than in the wild-type enzyme. There are two caveats, however; first, it is not certain that the differences in the observed rate constants are actually due to different rates of Compound I formation; second, a factor 3 in the rate constant translates to a rather small difference in free-energy barriers of 0.7 kcal/mol (much smaller than the differences of 4–9 kcal/mol in the rate-limiting QM/MM barriers for the wild-type enzyme and the Thr252MeO-Thr mutant).
We finally address the question of the preferred coupling mechanism in the Thr252MeO-Thr mutant. At face value, the rate-limiting barriers are somewhat lower in the Glu366 channel than in the Asp251 channel (Table 1). One should keep in mind, however, that the Asp251 channel is in contact with bulk water, so it should be rather facile to reprotonate Asp251 after each coupling reaction that involves proton transfer in the Asp251 channel. This is not true for Glu366, which resides in a hydrophobic pocket and is thus difficult to reprotonate. Protonation via the Asp251 channel may thus actually be a more realistic scenario for the coupling reaction, as in the case of the wild-type enzyme. In this scenario, the barrier for the initial homolytic cleavage is predicted to rise from 14.3 kcal/mol in the wild-type enzyme to 18.6 kcal/mol in the mutant. This increase in activation energy can be rationalized by an analysis of the hydrogen-bonding network. In the wild-type enzyme, the OH radical is stabilized in the transition state by hydrogen bonds to Thr252 and the FeO unit with distances of 1.64 and 2.06 Å, respectively [37]. Stabilization is less effective in the Thr252MeO-Thr mutant, where these distances increase to 2.06 and 2.22 Å, respectively (mechanism I in the Asp251 channel). The subsequent protonation, with concomitant electron transfer from the heme, is essentially downhill in the wild-type enzyme and requires some activation in the mutant (Table 1). This is not surprising, and gas-phase QM modeling of this process indeed confirms the qualitative expectation that the hydroxyl group in Thr252 is a better proton donor than the methoxy group in MeO-Thr252 (see Sect. 9 in the electronic supplementary material). In the mutant enzyme, this process is split into two steps (hydrogen transfer from the methoxy group to OH followed by a simultaneous proton and electron transfer in the Asp251 channel, see mechanism I) which make it energetically feasible, through the stabilization of the resulting intermediates by strong hydrogen bonds to two water molecules. Regardless of their limited quantitative accuracy, the present QM/MM results thus raise the possibility that residue 252 may play an active role in the proton delivery mechanism both for the wild-type enzyme and for the Thr252MeO-Thr mutant, whereas previous interpretations of the experimental data view this residue mainly as a structural factor for coordinating water molecules that deliver protons to the FeOOH unit [25].
We end with a cautionary note. The favored mechanism I in the Asp251 channel involves an incipient OH radical in the initial intermediate (IC1) that might be expected to undergo competing side reactions such as attack at the meso position of the porphyrin to affect heme degradation or demethylation of the methoxy group to regenerate Thr252. We note again in this context that the initially formed OH species is stabilized by hydrogen-bonding interactions with surrounding partners which lead to reduced OH spin density and hence presumably also to lower radical reactivity, in analogy to the situation in the wild-type enzyme [37]. A more reliable assessment will require QM/MM studies of the competing side reactions which are beyond the scope of this article.
References
Ortiz de Montellano PR (1998) Cytochrome P450: structure, mechanism and biochemistry. Plenum Press, New York
Ortiz de Montellano PR (1986) In: Ortiz de Montellano PR (ed) Cytochrome P450: structure, mechanism and biochemistry. Plenum Press, New York, pp 216–271
Dawson JH, Sono M (1987) Chem Rev 87:1255–1276
Sono M, Roach MP, Coulter ED, Dawson JH (1996) Chem Rev 96:2841–2888
Woggon WD (1997) Top Curr Chem 184:39–96
Ortiz de Montellano PR, De Voss JJ (2002) Nat Prod Rep 19:471–499
Watanabe Y (2001) J Biol Inorg Chem 8:846–856
Denisov IG, Makris TM, Sligar SG, Schlichting I (2005) Chem Rev 105:2253–2278
Schröder D, Schwarz H, Shaik S (2000) Struct Bond 97:91–123
Groves JT (2000) J Porphyrins Phthalocyanines 4:350–352
Meunier B (2000) J Porphyrins Phthalocyanines 4:353
Woggon WD, Wagenknecht HA, Claude C (2001) J Inorg Biochem 83:289–300
Costas M, Mehn MP, Jensen MP, Que L Jr (2004) Chem Rev 104:939–986
Rohde JU, In JH, Lim MH, Brennessel WW, Bukowski MR, Stubna A, Münck E, Nam W, Que L Jr (2003) Science 299:1037–1039
Imai M, Shimada H, Watanabe Y, Matsuhima-Hibiya Y, Makino R, Koga H, Horiuchi T, Ishimura Y (1989) Proc Natl Acad Sci USA 86:7823–7827
Martinis SA, Atkins WM, Stayton PS, Sligar SG (1989) J Am Chem Soc 111:9252–9253
Gerber NC, Sligar SG (1994) J Biol Chem 269:4260–4266
Gerber NC, Sligar SG (1992) J Am Chem Soc 114:8742–8743
Loew GH, Rohmer MM (1980) J Am Chem Soc 102:3655–3657
Poulos TL, Anthony-Cahill SJ, Griffith MC, Schultz PG (1989) Science 244:182–188
Harris DL, Loew GH (1994) J Am Chem Soc 116:11671–11674
Makris TM, Denisov IG, Sligar SG (2003) Biochem Soc Trans 31:516–519
Davydov R, Kappl R, Huttermann J, Peterson JA (1991) FEBS Lett 295:113–115
Davydov R, Macdonald IDG, Makris TM, Sligar SG, Hoffman BM (1999) J Am Chem Soc 121:10654–10655
Kimata Y, Shimada H, Hirose T, Ishimura Y (1995) Biochem Biophys Res Commun 208:96–102
Newcomb M, Toy PH (2000) Acc Chem Res 33:449–455
Vaz ADN, McGinnity DF, Coon MJ (1998) Proc Natl Acad Sci USA 95:3555–3560
Harris DL, Loew GH (1998) J Am Chem Soc 120:8941–8948
Guallar V, Friesner RA (2004) J Am Chem Soc 126:8501–8508
Kamachi T, Shiota Y, Ohta T, Yoshizawa K (2003) Bull Chem Soc Jpn 76:721–732
Taraphder S, Hummer G (2003) J Am Chem Soc 125:3931–3940
Kumar D, Hirao H, de Visser SP, Zheng J, Wang D, Thiel W, Shaik S (2005) J Phys Chem B 109:19946–19951
Shaik S, Kumar D, de Visser SP, Altun A, Thiel W (2005) Chem Rev 105:2279–2328
Ogliaro F, de Visser SP, Cohen S, Sharma PK, Shaik S (2002) J Am Chem Soc 124:2806–2817
Derat E, Shaik S (2006) J Phys Chem B 110:10526–10533
Sen K, Hackett JC (2009) J Phys Chem B 113:8170–8182
Zheng J, Wang D, Thiel W, Shaik S (2006) J Am Chem Soc 128:13204–13215
Chen H, Moreau Y, Derat E, Shaik S (2008) J Am Chem Soc 130:1953–1965
Chen H, Hirao H, Derat E, Schlichting I, Shaik S (2008) J Phys Chem B 112:9490–9500
Altarsha M, Benighaus T, Kumar D, Thiel W (2009) J Am Chem Soc 131:4755–4763
Schöneboom JC, Lin H, Reuter N, Thiel W, Cohen S, Ogliaro F, Shaik S (2002) J Am Chem Soc 124:8142–8151
Schöneboom JC, Thiel W (2004) J Phys Chem B 108:7468–7478
Altun A, Thiel W (2005) J Phys Chem B 109:1268–1280
Vidakovic M, Sligar SG, Li H, Poulos TL (1998) Biochemistry 37:9211–9219
Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML (1983) J Chem Phys 79:926–935
MacKerell AD Jr, Bashford D, Bellott M, Dunbrack RL Jr, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE III, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiórkiewicz-Kuczera J, Yin D, Karplus M (1998) J Phys Chem B 102:3586–3616
Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M (1983) J Comput Chem 4:187–217
Becke AD (1993) J Chem Phys 98:5648–5652
Hay PJ, Wadt WR (1985) J Chem Phys 82:299–310
Hehre WJ, Ditchfield K, Pople JA (1972) J Chem Phys 56:2257–2261
Schäfer A, Horn H, Ahlrichs R (1992) J Chem Phys 97:2571–2577
Schäfer A, Huber C, Ahlrichs R (1994) J Chem Phys 100:5829–5835
Bakowies D, Thiel W (1996) J Phys Chem 100:10580–10594
Ahlrichs R, Bär M, Häser M, Horn H, Kölmel C (1989) Chem Phys Lett 162:165–169
Smith W, Forester TR (1996) J Mol Graph 14:136–141
Sherwood P, De Vries AH, Guest MF, Schreckenbach G, Catlow CRA, French SA, Sokol AA, Bromley ST, Thiel W, Turner AJ, Billeter S, Terstegen F, Thiel S, Kendrick J, Rogers SC, Casci J, Watson M, King F, Karlsen E, Sjøvoll M, Fahmi A, Schäfer A, Lennartz C (2003) J Mol Struct 632:1–28
Billeter SR, Turner AJ, Thiel W (2000) Phys Chem Chem Phys 2:2177–2186
Siegbahn PEM, Blomberg MRA, Crabtree RH (1997) Theor Chem Acc 97:289–300
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Altarsha, M., Benighaus, T., Kumar, D. et al. Coupling and uncoupling mechanisms in the methoxythreonine mutant of cytochrome P450cam: a quantum mechanical/molecular mechanical study. J Biol Inorg Chem 15, 361–372 (2010). https://doi.org/10.1007/s00775-009-0608-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00775-009-0608-3