Abstract
Employing site-directed spin labeling (SDSL), the structure of maltose-binding protein (MBP) had previously been studied in the native state by electron paramagnetic resonance (EPR) spectroscopy. Several spin-labeled double cysteine mutants were distributed all over the structure of this cysteine-free protein and revealed distance information between the nitroxide residues from double electron–electron resonance (DEER). The results were in good agreement with the known X-ray structure. We have now extended these studies to the molten globule (MG) state, a folding intermediate, which can be stabilized around pH 3 and that is characterized by secondary but hardly any tertiary structure. Instead of clearly defined distance features as found in the native state, several additional characteristics indicate that the MG structure of MBP contains different polypeptide chain and domain orientations. MBP is also known to bind its substrate maltose even in MG state although with lower affinity. Additionally, we have now created new mutants allowing for spin labeling at or near the active site. Our data confirm an already preformed ligand site structure in the MG explaining its substrate binding capability and thus most probably serving as a nucleation center for the final native structure.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Maltose-binding protein (MBP) in E. coli is a single-chain polypeptide of 370 amino acid residues devoid of cofactors. It is composed of two domains with the active site located in a cleft between the domains primarily binding maltose. Substrate binding is accompanied by a conformational change of the protein from an open to a closed state, with the two domains getting closer together. The native apoprotein is devoid of any cysteines [1]. In its physiological context, it is an important part of the maltose translocation machinery. Thus, earlier CW EPR studies at about physiological pH were already studying the spectral changes upon maltose binding [2,3,4] that are indicative of slight structural rearrangements. In a previous work, we have employed seven spin-labeled cysteine mutants (MBP1 to MBP7) that are distributed over the whole structure for shape triangulation. The distances between the biradicals in single pairs were determined by double electron–electron resonance spectroscopy (DEER) and found to be in excellent agreement with theoretical predictions from molecular modeling of the known X-ray structure (Fig. 1) [5].

reproduced from Reichenwallner et al. [5] with kind permission of the publisher
Structural representation of spin-labeled mutants of MBP (a) and their relative orientation and distance to the P298C residue located in the center of the maltose-binding active site. Figure
Further to this, we could follow the maltose-dependent opening and closing of the protein with an additional set of spin-labeled mutants close to the active site employing DQC (double quantum coherence) EPR spectroscopy [6].
The molten globule (MG) state is an intensively studied intermediate in protein folding and is defined by ANS binding due to the increased exposure of hydrophobic patches [7,8,9]. However, it contains substantial amounts of secondary structure but lacks the tight and precise packing typical of the native state. At pH values around 3, this state can be stabilized in case of MBP. It has been shown that MBP still binds maltose under these acidic conditions, although rather weakly [10].
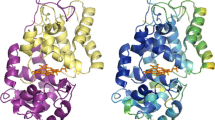
In this study, we conducted four-pulse DEER experiments [11, 12] to determine the interspin distances of these seven mutants of MBP, now in the MG state. Based on the findings of these experiments, we also aimed to determine if there is already a preformed structure around the binding pocket of MBP in the molten globule as it does bind the substrate maltose under those conditions. To enable this investigation, another two double mutants MBP8 (S233C P298C) and MBP12 (G13C T237C) were constructed. The mutated cysteine pairs for the attachment of spin labels are located at or near the binding pocket as shown in the MBP-containing crystal structure (Fig. 2, PDB ID: 1anf) [13] and distances were determined with continuous wave (CW) EPR (Fig. 2) [14, 15]. Moreover, the binding affinity of maltose towards MBP in the MG state was determined by ITC.
Structural representation of MBP8 and MBP12 with Discovery Studio Visualizer BIOVIA (PDB ID: 1anf, modified). The mutated cysteines are displayed in a stick representation. The respective cysteine pairs are connected by blue lines and are indicated with corresponding mutant names (MBP8 is S233C P298C and MBP12 is G13C T237C). In the center of the picture, the maltose molecule is also given in stick representation. The perspective is from one side with the two domains displayed in top right and bottom left (colour figure online)
1.1 Materials and Methods
1.1.1 Materials
All reagents were from commercial sources with analytical grade. (1-Oxyl-2,2,5,5-tetramethyl-pyrroline-3-methyl) methanethiosulfonate (MTS) was a kind gift from Kálmán Hideg (University of Pécs, Pécs, Hungary).
Plasmids of the mutants MBP1 to MBP7 are described in [5]. The plasmids of the double mutants MBP8 (S233C P298C) and MBP12 (G13C T237C) were mutated in such a way that each pair of cysteines is located near the binding pocket.
1.1.2 Protein Purification, Spin Labeling and Sample Preparation
The protein samples for DEER experiments were purified and spin-labeled as described previously [5] to generate comparable data. The protein samples for CW EPR were dialyzed for desalting and buffer exchanges instead of using a PD-10 column. The residual content of maltose was determined by a coupled maltose assay (Sigma-Aldrich) based on enzymatic reactions, in which maltose is degraded to glucose (α-glucosidase), and further phosphorylated (hexokinase) and dehydrogenated (glucose-6-phosphate dehydrogenase) to 6-phosphogluconate [16]. CGH5 (citrate, glycine, and HEPES, 5 mM each) buffer was used for all of the samples.
The protein samples used for the MG study were adjusted to pH 3 [17] via dialysis. The samples used for CW EPR and DEER experiments contained 0.15 mM of spin-labeled protein, 20% (v/v) glycerol, and 17.2 mM maltose if required. Samples containing maltose were incubated for at least 5 min. All samples were shock frozen and stored at − 80 °C until use.
1.1.3 Experiment
ITC experiments were performed at 5 °C. Protein samples were examined at pH 3.2 and pH 7.5. For either pH value, experiments were performed in triplicate. The concentrations of MBP wildtype (MBPwt) and maltose were 0.05 mM and 0.5 mM at pH 7.5, respectively, while at pH 3.2, the concentrations of MBPwt and maltose were 0.5 mM and 2.5 mM for the first trial, and 0.39 mM and 1.17 mM for the last two trials, respectively. The heats of injection were integrated with NITPIC [18] and further globally fitted with SEDPHAT [19].
1.1.4 EPR Experiments
Four-pulse DEER experiments were performed using the sequence [11, 12]:
to obtain dipolar time evolution data. All pulse EPR experiments were performed at X-band frequencies of 9.24–9.37 GHz using BRUKER Elexsys E580 spectrometers, each equipped with BRUKER Flexline split-ring resonators ER4118X–MS3 (BRUKER, Billerica, MA, USA). The temperatures were set to 50 K for all experiments. The resonators were overcoupled to Q ≈ 200. The pump frequency νpump was set to the maximum of the field-swept electron spin echo. The observer frequency νobs was set to νpump + 65 MHz. The observer pulse lengths for each DEER experiment were set to 32 ns for both π/2- and π-pulses and the pump pulse length was 12 ns.
The low-temperature CW EPR experiments were also performed on a BRUKER Elexsys E580 with a dielectric resonator (BRUKER Flexline ER 4118X MD5) at 150 K, a frequency of about 9.78 GHz, a microwave power of 6.4 mW and a modulation amplitude of 1 G. For low-temperature setups (150 K), the microwave power was lowered to 0.064 mW. The dipolar broadened CW spectra were fitted with DIPFIT 2.0 based on nonlinear least-squares fitting of simulated EPR spectra to experimental spectra [15], while the DEER time domain signals were fitted with DeerAnalysis 2011 and MATLAB R2013b using Tikhonov regularization [20].
2 Results and Discussion
2.1 MBP1 to MBP7 Represent the Global Structure of the Two Domains
Here, interspin distances of the spin-labeled mutants MBP1 to MBP7 were determined in their molten globule states at pH 3 as it was previously found by ANS binding [5, 10]. Figure 3a illustrates the time domain signals, and the associated distance distributions of the seven MBP mutants at pH 3 are shown in Fig. 3b (published in part in the doctoral thesis of Mohammed Chakour [21]). For example, MBP5 exhibits multiple distance characteristics at 2.3, 2.8 and 3.8 nm at pH 3, while the distance is unique at about 1.8 nm at pH 7 as shown in previous work [5]. The distance distribution at pH 3 is explicitly broadened and more peaks appear as compared to pH 7. MBP1, 2, 3, 4 and 7 exhibit the same tendency. MBP6 does not considerably change between native and MG state.
DEER experiments on MBP1 to MBP7 at pH 3. a The time domain signals and (b) distance distributions from Tikhonov regularization. Apart from MBP6, the distance distributions of the spin-labeled mutants are broadened in the MG state compared to pH 7 [5]
Furthermore, in the presence of excess maltose, MBP5 yields the distances of 2.4, 3.1 and 3.9 nm at pH 3. Although individual distances appear similar, the population density of the peaks is slightly altered with a preference to shorter distances (Fig. 4). MBP7 behaves similarly as MBP5 in the presence and absence of maltose at pH 3.
2.2 Binding Affinity of Maltose to MBP in the MG State is Confirmed by ITC
DEER data of MBP5 and MBP7 at pH 3 indicate that MBP still interacts with maltose at pH 3. To confirm this binding event at pH 3, ITC experiments were performed. For comparison, a measurement at pH 7 was carried out as well; the results are shown in Fig. 5 and Table 1. Each single experiment was processed with NITPIC, and integrated signals extracted from NITPIC of three independent experiments at each pH value were fitted globally with SEDPHAT.
The dissociation constant at pH 3.2 is 28.8 µM, which is 11-fold larger than at pH 7.5 (2.6 µM). This indicates that MBP in the native state has a higher affinity to maltose than in the MG state, which corroborates the results of Varadarajan and co-workers [10]. Nevertheless, it is evident from these results that MBP is still capable of binding its natural substrate under acidic, strongly destabilizing conditions that favor population of the MG.
All experiments were performed at 5 °C. Figures in parentheses indicate the confidence interval with a confidence level of 95%.
2.3 MBP8 and MBP12 Represent the Structure Near the Binding Pocket
After confirming that maltose binds to MBP at pH 3.2 and thus affects its peripheral tertiary structure, it is of interest to probe the structure variation around the binding pocket in the MG state. The mutants MBP8 and MBP12 were constructed such that they possess the mutated cysteine pairs near the binding pocket. The resulting short distances for MBP8 and MBP12 can be investigated by low-temperature CW EPR as shown in Fig. 6. The samples were prepared in the presence and absence of maltose, both at pH 3 and pH 7.
The mean distances and widths of the distance distributions as calculated with DIPFIT [15] from the spectra above are listed in Table 2. At pH 3 MBP8 and MBP12 present almost the same distance and width, respectively, regardless of the presence of maltose. Although MBP8 presents a width at pH 7 smaller than at pH 3, the mean distances are comparable at both pH values. MBP12 shows only little difference between pH 3 and pH 7, even in the absence of maltose.
For MBP8, a slight structural contraction is observed at pH 7 upon binding to maltose. However, the resulting distribution widths are quite broad in all cases.
3 Conclusion
The mutants MBP1 to MBP7 reveal changes in the global tertiary structure of MBP, which stretch across both domains. In the MG state of MBP, the structure is undefined in most regions of MBP in view of the broadened distance distributions and the increased number of distance peaks within MBP1 to MBP7. In the presence of excess maltose, the distance distributions in the MG state differ from those in the absence of maltose, which implies that MBP binds to maltose in the MG state and a rather undefined additional conformational change occurs. The binding process of MBP to maltose in the MG state was confirmed by ITC experiments, although exhibiting an 11-fold lower affinity than in the native state. Unfortunately, such experiments are not feasible with the mutants due to the lower affinity. This process (at 5 °C) is enthalpically less unfavorable with ΔH of 2.6 kcal/mol in the molten globule state than 3.8 kcal/mole in the native state, which might indicate more facile ligand binding to the MG than to the native structure. Entropically, ligand binding is favored in the native state as compared to the MG state, suggesting that larger structural rearrangements of the polypeptide chain need to occur upon ligand binding to the MG. Maltose binding thus partially stabilizes the tertiary structure of MBP. These results agree well with those of Varadarajan and co-workers for MBP [10]. Hilvert and co-workers found a similar behavior for monomeric chorismate dismutase (mMjCM) that constitutes a catalytic active molten globule [22, 23].
Ligands are generally well known to stabilize tertiary structures proteins as, e.g., for fatty acid binding to human serum albumin [24]. Similar EPR-based findings were also made for other proteins, like intrinsically disordered proteins (IDPs), or membrane proteins that stabilize upon ligand binding, as it was recently discussed by Jeschke as a ‘fuzzy relation of structure to function’ [25].
For MBP, the question arises whether maltose induces the tertiary structure from random structures, or if the binding pocket is already pre-formed and maltose, therefore, binds to it and helps stabilizing the rest of the tertiary structure. With mutants MBP8 and MBP12, we had hoped to answer this question. There are, however, no clear differences of the mean distances at both pH values based on the low-temperature CW EPR spectra of MBP8 and MBP12, besides the broadened width in case of MBP8 (Fig. 6). Furthermore, maltose binding does not lead to a significant change of distance distributions in the native versus the MG state. This suggests the presence of a defined structure near the binding pocket of MBP in the MG state again confirming the assumption of [10]. However, DIPFIT can only give an average distance with line shapes that only weakly depend on the actual distance. Low-temperature CW EPR is restricted to a quite narrow distance range (~ 8 to 20 Å) and in general, the uncertainties in the calculated distances from CW EPR can be substantial. Unfortunately, these distances are too small to be detected in DEER experiments that usually provide information from 18 to 60 Å and under favorable conditions up to even 15–160 Å [25, 26]. Additional double quantum coherence (DQC) experiments may yield more precise distance information from MBP8 and MBP12 as was recently shown [6].
All the evidence above leads to the conclusion that MBP maintains a native-like structure around the binding pocket in the MG state, an obviously essential prerequisite for binding of maltose. In turn, maltose binding stabilizes the rest of the molecule. These findings strongly support recent trends in structural biology that suggest reconsidering some general aspects in protein folding and function [25].
Abbreviations
- ANS:
-
8-Anilinonaphthalene-1-sulfonic acid
- CW:
-
Continuous wave
- DEER:
-
Double electron–electron resonance
- DQC:
-
Double quantum coherence
- EPR:
-
Electron paramagnetic resonance
- IDP:
-
Intrinsically disordered protein
- ITC:
-
Isothermal titration calorimetry
- MBP:
-
Maltose-binding protein
- MG:
-
Molten globule
- MTS:
-
(1-Oxyl-2,2,5,5-tetramethyl-pyrroline-3-methyl) methanethiosulfonate
- SDSL:
-
Site-directed spin labeling
References
J.C. Spurlino, G.Y. Lu, F.A. Quiocho, J. Biol. Chem. 266(8), 5202–5219 (1991)
J.A. Hall, T.E. Thorgeirsson, J. Liu, Y.K. Shin, H. Nikaido, J. Biol. Chem. (1997). https://doi.org/10.1074/jbc.272.28.17610
M.I. Austermuhle, J.A. Hall, C.S. Klug, A.L. Davidson, J. Biol. Chem. (2004). https://doi.org/10.1074/jbc.M403508200
J. Cui, S. Qasim, A.L. Davidson, J. Biol. Chem. (2010). https://doi.org/10.1074/jbc.M110.147819
J. Reichenwallner, M. Chakour, S. Indu, R. Varadarajan, W.E. Trommer, Appl. Magn. Reson. (2013) doi: 10.1007/s00723-013-0468-4
B. Selmke, P.P. Borbat, C. Nickolaus, R. Varadarajan, J.H. Freed, W.E. Trommer, Biochemistry (2018). https://doi.org/10.1021/acs.biochem.8b00322
D.A. Dolgikh, R.I. Gilmanshin, E.V. Brazhnikov, V.E. Bychkova, G.V. Semisotnov, S.Y. Venyaminov, O.B. Ptitsyn, FEBS Lett. (1981). https://doi.org/10.1016/0014-5793(81)80642-4
O.B. Ptitsyn, V.N. Uversky, FEBS Lett. (1994). https://doi.org/10.1016/0014-5793(94)80231-9
J. Reichenwallner, M.T. Oehmichen, C.E.H. Schmelzer, T. Hauenschild, A. Kerth, D. Hinderberger, Magnetochemistry (2018). https://doi.org/10.3390/magnetochemistry4040047
R.S. Prajapati, S. Indu, R. Varadarajan, Biochemistry (2007). https://doi.org/10.1021/bi700577m
R.E. Martin, M. Pannier, F. Diederich, V. Gramlich, M. Hubrich, H.W. Spiess, Angew. Chem. (1998). https://doi.org/10.1002/(SICI)1521-3773(19981102)37:20<2833:AID-ANIE2833>3.0.CO;2-7
M. Pannier, S. Veit, A. Godt, G. Jeschke, H.W. Spiess (2011) J. Magn. Reson. (San Diego, Calif: 1997). https://doi.org/10.1016/j.jmr.2011.08.035
F.A. Quiocho, J.C. Spurlino, L.E. Rodseth, Structure (1997). https://doi.org/10.1016/S0969-2126(97)00253-0
M.D. Rabenstein, Y.K. Shin, Proc. Natl. Acad. Sci. U.S.A. (1995). https://doi.org/10.1073/pnas.92.18.8239
H.J. Steinhoff, N. Radzwill, W. Thevis, V. Lenz, D. Brandenburg, A. Antson, G. Dodson, A. Wollmer, Biophys. J. (1997). https://doi.org/10.1016/S0006-3495(97)78353-X
H.U. Bergmeyer (ed), Methoden der enzymatischen Analyse, 3rd edn. (Verl. Chemie, Weinheim/Bergstr., 1974)
S. Sheshadri, G.M. Lingaraju, R. Varadarajan, Protein Sci. (1999). https://doi.org/10.1110/ps.8.8.1689
S. Keller, C. Vargas, H. Zhao, G. Piszczek, C.A. Brautigam, P. Schuck, Anal. Chem. (2012). https://doi.org/10.1021/ac3007522
P. Schuck, Biophys. J. (2000). https://doi.org/10.1016/S0006-3495(00)76713-0
G. Jeschke, V. Chechik, P. Ionita, A. Godt, H. Zimmermann, J. Banham, C.R. Timmel, D. Hilger, H. Jung, Appl. Magn. Reson. (2006) doi: 10.1007/BF03166213
M. Chakour, Strukturuntersuchungen verschiedener Mutanten des Maltose-Bindungsproteins (MBP) mit Hilfe der ESR Spektroskopie. Doctoralthesis (Technische Universität Kaiserslautern, 2015)
K. Vamvaca, B. Vögeli, P. Kast, K. Pervushin, D. Hilvert, Proc. Natl. Acad. Sci. U.S.A. (2004). https://doi.org/10.1073/pnas.0404109101
K. Vamvaca, I. Jelesarov, D. Hilvert, J. Mol. Biol. (2008). https://doi.org/10.1016/j.jmb.2008.07.049
A. Shrake, D. Frazier, F.P. Schwarz, Biopolymers (2006). https://doi.org/10.1002/bip.20406
G. Jeschke, Emerging Topics Life Sciences (2018). https://doi.org/10.1042/ETLS20170143
T. Schmidt, M.A. Wälti, J.L. Baber, E.J. Hustedt, G.M. Clore, Angew. Chem. Int. Ed. (2016). https://doi.org/10.1002/anie.201609617
Funding
Open Access funding enabled and organized by Projekt DEAL. Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Nickolaus, C., Vargas, C., Reichenwallner, J. et al. The Molten Globule State of Maltose-Binding Protein: Structural and Thermodynamic Characterization by EPR Spectroscopy and Isothermal Titration Calorimetry. Appl Magn Reson 51, 877–886 (2020). https://doi.org/10.1007/s00723-020-01232-y
Received:
Revised:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00723-020-01232-y