Abstract
ARFs in plants mediate auxin signaling transduction and regulate growth process. To determine genome-wide characterization of ARFs family in melon (Cucumis melo L.), ARFs were identified via analysis of information within the melon genomic database, and bioinformatic analyses were performed using various types of software. Based on different treatment methods involving dipping with the growth regulator Fengchanji No. 2 and artificial pollination, Jingmi No. 11 melon was used as the test material, and melon plants with unpollinated ovaries served as controls. The expression of ARFs during the early development of melon was analyzed via qRT-PCR. Seventeen genes that encode ARF proteins were identified in the melon genome for the first time. The expression of these ARFs differed in different tissues. The expression levels of CmARF2, CmARF16-like, CmARF18-like2, and CmARF19-like were especially high in melon fruits. The expression of ARFs during the early development of melon fruits differed in response to the different treatments, which suggested that CmARF9, CmARF16-like, CmARF19-like, CmARF19, CmARF1, CmARF2, CmARF3, and CmARF5 may be associated with melon fruit growth during early development. Interestingly, the increase in the transverse diameter of fruits treated with growth regulators was significantly greater than that of fruits resulting from artificial pollination, while the increase in the longitudinal diameter of the fruits resulting from artificial pollination was significantly greater.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Melon (Cucumis melo L.) is an important horticultural crop species that is cultivated in temperate, subtropical, and tropical regions worldwide. Auxin is a plant hormone that plays pivotal roles in the regulation of plant growth in response to various developmental and environmental events, such as embryogenesis, organogenesis, trophic growth, root architecture development, flower and fruit development, tissue and organ patterning, and vascular development (Davies 1987; Zhang et al. 2012; Vanneste and Friml 2009). It has been shown that auxin coordinates plant development essentially via the transcriptional regulation of members of certain gene families, such as the auxin/indole-3-acetic acid (Aux/IAA), Gretchen Hagen 3 (GH3), small auxin-up RNA (SAUR), and auxin response factor (ARF) families (Tiwari et al. 2001; Park et al. 2007; ElsharkawyI et al. 2014). Recent studies have shown that auxin signal transduction may be accomplished via the regulation of ARFs. These so-called early auxin-responsive genes are characterized by conserved promoter elements, such as the TGA element (AACGAC), core element of the auxin response region (AuxRE-core; GGTCCAT), and auxin response element (AuxRE; TGTCTC) (Guilfoyle and Hagen 2007; Tiwari et al. 2003). As important components of the auxin signaling pathway, ARFs activate or repress the expression of auxin-responsive genes by binding to AuxREs in the promoters of those genes (Ulmasov et al. 1999).
A typical ARF consists of a highly conserved N-terminal B3-like DNA-binding domain (DBD) that recognizes AuxREs in the promoters of auxin-responsive genes and a C-terminal dimerization domain (CTD) that contains two motifs, namely, III and IV, which are also found in Aux/IAA proteins and enable the formation of homodimers and heterodimers between ARFs and Aux/IAAs (Ulmasov et al. 1999). The middle region (MR), which is located between the DBD and CTD, can perform transcriptional activation or repression depending on the amino acid composition (Ellis et al. 2005). Analysis of the amino acid compositions of activation dimers (ADs) and repression dimers (RDs) of ARFs revealed that ADs are rich in glutamine (Q), serine (S), and leucine (L) residues, while RDs are rich in proline (P), serine (S), threonine (T), and glycine (G) residues (Guilfoyle and Hagen 2007; Tiwari et al. 2003; Ulmasov et al. 1999; Ellis et al. 2005).
ARF proteins participate in the transcriptional regulation of a variety of biological processes associated with growth and development, such as embryogenesis (Thorsten et al. 2002; Dolf et al. 2014), leaf expansion (Wilmoth et al. 2010), leaf senescence (Lim et al. 2010), lateral root growth (Tatematsu et al. 2004; Yoko et al. 2007), and fruit development (Marc et al. 2006; Goetz et al. 2007; et al. 2010). The genome sequencing project for melon was recently completed (Garcia-Mas et al. 2012), and the ARFs of Arabidopsis (Piya et al. 2014), Oryza (Wang et al. 2007), Populus (Yang et al. 2014), Medicago (Shen et al. 2015), and maize (Liu et al. 2011) have also been published. However, genome-wide information concerning the ARF gene family and the functions of ARFs in melon are largely unknown. Therefore, to improve any fruit-related agronomic traits in melon, it is necessary to study the characteristics of members of the ARF family and their potential functions during early fruit development in melon.
To elucidate the structures of CmARFs and characterize the expression of CmARFs during fruit development in melon, 17 putative genes that contain ARF domains were identified via genomic data mining. The sequence homology of all the CmARFs was then investigated, followed by a comparative phylogenetic analysis. The different temporal and spatial expression patterns during early fruit development of melon were subsequently determined for each CmARF gene by quantitative real-time PCR (qRT-PCR). The resulting classification of groups and characterization of the expression patterns will be useful for future analyses of the biological functions of ARF family genes in melon.
Materials and methods
Plant material and treatments
Seeds of melon (Cucumis melo L.) cultivar Jingmi No. 11, a popular oriental melon variety provided by the Beijing Agricultural Technology Promotion Station, were sown in an organic matter:soil (2:1 v/v) mixture at Beijing the University of Agriculture Experimental Station. The plants were cultivated under glasshouse conditions (12 h of light [300~1300 μmol m−2 s−1 light] and 26 ± 2 °C during the day; 12 h of darkness and 15 ± 2 °C during the night; 50~70% relative humidity) and were watered and fertilized in accordance with normal cultivation management practices. Female flowers were artificially pollinated or were subjected to dipping with the growth regulator Fengchanji No. 2 (a hormone complex that contains mainly 4-chlorophenoxyacetic acid to increase the rate of fruit set; Shenyang Agricultural University), and plants with unpollinated ovaries served as controls. Female flowers before 1 day of treatment were bagged to avoid pollination. The first day of blooming was considered day 0, and those plants were tagged. Plants were trained as single stems, and the fruit load was limited to three per lateral vine per plant from the 12th to 14th nodes.
Fruits were collected at 0, 1, 2, 3, and 5 days after treatment. Some of the fruit samples were used to analyze growth, and the transverse diameters and longitudinal diameters (in cm) of the fruits from the control and treated plants were measured with a Vernier caliper. The other collected fruit samples from the control and treated plants were frozen in liquid nitrogen and stored at − 80 °C until further gene expression analysis. Moreover, leaf, pistillate flower, staminate flower, stem, and root tissues from the control and treated plants were also collected for tissue-specific expression analysis.
Identification of melon ARF genes
To find previously identified and potential ARF family genes in melon, multiple database searches were performed. The key term “auxin response factor” was used to query the melon database (http://melonomics.net). After searching for ARF genes, we also queried the database via the amino acid sequences of the ARF domains of several known ARF family members (including AtARFs and OsARFs). To confirm the accuracy of the gene identification further, the predicted CmARF-like gene sequences were compared with the sequences of ARF proteins from other species via BLASTp retrieval. Only those sequences with high scores (> 200) were selected.
Sequence analysis
ProtParam (Mcphie 2002) (http://web.expasy.org/protparam) was used to analyze the protein parameters. The neighbor-joining (NJ) algorithm of the MEGA 6.0 program (Tamura et al. 2011) was used to construct a phylogenetic tree with Poisson correction and pairwise deletion parameters, and all other parameters were configured to the default values. A total of 1000 bootstrap replicates were included. The subcellular localization of the deduced polypeptides was predicted by WoLF PSORT (https://wolfpsort.hgc.jp/). Moreover, GSDS (Guo et al. 2007) (http:/gsds.cbi.pku.edu.cn/) was used to map the exon-intron structure of the ARF family members, and the conserved domains of the proteins were predicted using MEME Suite (http://meme-suite.org/tools/meme) (Bailey et al. 2009).
RNA isolation and cDNA synthesis
Total RNA was isolated using TransZol (TransGen Biotech, Beijing, China) according to the manufacturer’s instructions. The RNA integrity was determined by electrophoresis on 1% agarose gels. The quantity and purity of the RNA were determined using a NanoDrop™ 2000 spectrophotometer (Thermo Fisher Scientific, Massachusetts, United States). Only high-quality samples in which A260:A280 ≥ 1.8 and A260:/A230 ≥ 2.0 were used for subsequent cDNA synthesis. cDNA was synthesized using a Prime-Script RT Reagent Kit (Perfect Real Time, TaKaRa, Tokyo, Japan) according to the manufacturer’s instructions. For each sample, 1 mg of total RNA was used for each 20-mL reverse transcription reaction system.
Semiquantitative PCR and qRT-PCR
For semiquantitative PCR and qRT-PCR, gene-specific oligonucleotide primers for each ARF gene were designed by Beacon Designer; these primers are listed in Supplemental Table 1. The specificity of each pair of primers was determined by agarose gel electrophoresis and by resequencing the PCR products. Twenty cycles were carried out for the CmARFs.
The reaction system (10 μL) contained 2 × SYBR qPCR mix (5 μL), cDNA template (1 μL), 10 μM forward primer (0.5 μL), 10 μM reverse primer (0.5 μL), and ddH2O (3 μL). The reaction procedure was as follows: 3 min of predenaturation at 95 °C; 40 cycles of 10 s at 94 °C, 30 s of annealing (the annealing temperature was set according to the information in Supplemental Table 1), 20 s at 72 °C, and 5 min of extension at 72 °C (Weng et al. 2017). The 2−ΔΔCT relative quantitation method was then used to calculate the relative expression of the ARF genes (Livak and Schmittgen 2001). The 18S rRNA gene (148 bp) of melon was used as an internal control. A Bio-Rad CFX 96 real-time PCR instrument and CFX manager 3.0 (Bio-Rad Lab., CA, USA) were used for real-time PCR analysis. Each experiment was repeated at least three times.
Statistical analysis
The experiment was performed in triplicate. With respect to the measurements of the transverse diameters and longitudinal diameters (cm) of the fruits, each biological replicate consisted of five samples from five individual plants. With respect to the gene expression analyses, three different fruits were pooled together as one biological sample, and this was performed three times to produce three independent biological replicates (each consisting of three pooled fruits). The data represent the means ± SDs of three replications and were statistically analyzed using analysis of variance (ANOVA) by SPSS 10.0 (International Business Machine, Chicago, IL, USA). Tukey’s test was subsequently used to determine significant differences among the groups (p < 0.05, p < 0.01). Figures representing the morphological and gene expression parameters were drawn using Origin Pro 8.0 SR4 (Origin Lab, Northampton, MA, USA).
Results
Identification and bioinformatic analyses of melon ARF genes
The sequences of all the predicted ARF genes in the melon genome were compiled and compared with those of ARF genes from other species. Seventeen ARF genes with complete structural domains were ultimately identified in melon. The CmARFs were named on the basis of the results of the comparison with the other species. Three of the ARF18-like genes were distinguished by adding one number to the end.
Using ExPASy, we found that the longest ARF protein consists of 1110 amino acid residues, while the shortest ARF protein consists of 584 amino acid residues. The ORF length ranges from 1752 to 3330 bp. In addition, the isoelectric points of the 17 ARF proteins were also predicted to range from 5.42 to 8.67, as shown in Table 1. Via the WoLF PSORT program, 16 ARF proteins were determined to be located in the nucleus, whereas CmARF5 was predicted to be located in the peroxisome.
The amino acid sequences of the CmARFs were analyzed by MEME software for the selection of 8 conserved sequences, as shown in Fig. 1. Motifs 1 and 2 were located in the structural DBD of the CmARFs, and motifs 5, 6, and 7 were located in the MR conserved structural domain of the CmARFs. These findings indicate that CmARFs have a highly conserved motif in both the DBD and the MR domain.
Analysis of gene structure and phylogenetic relationships
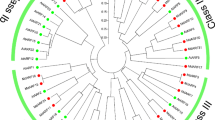
Phylogenetic analysis involving the deduced amino acid sequences of the 17 melon ARFs and 21 ARFs from Arabidopsis thaliana was performed. The results showed that the ARF family genes of the two species could be divided into four subfamilies: classes I, II, III, and IV (Fig. 2). ARFs exhibit high homology between melon and Arabidopsis thaliana.
Phylogenetic relationship of putative ARFs in Arabidopsis thaliana and melon. The phylogenetic tree was constructed using the NJ method with MEGA 7.0 software. Bootstrap values (1000 replicates) are shown as percentages at the branch nodes. Arabic numerals I–IV represent different ARF groups. The results indicated that ARFs exhibit high homology between melon and Arabidopsis thaliana
The phylogenetic tree revealed the following information: there are also four subfamilies of ARFs in melon, classes I, II, III, and IV (Fig. 3a). Structural analysis of the identified melon ARF genes revealed differences in the numbers and locations of introns. Most members of the ARF gene family have a complex structure, with approximately 10 introns. There were fewer than five introns in only four ARF genes, namely, CmARF16-like, CmARF17, CmARF18, and CmARF18-like1 (Fig. 3b). Most CmARFs contained three typical domains: a DBD, an auxin_resp domain, and a CTD. CmARF3, CmARF16-like, CmARF17, CmARF18, and CmARF18-like1 contained a DBD and an auxin_resp domain but no CTD (Fig. 4).
Phylogenetic analysis and intron/exon structures of putative ARF genes in melon. a. Phylogenetic analysis of putative ARF genes in melon. ClustalW was used to align the protein sequences of 17 CmARFs. The phylogenetic tree was constructed using the NJ method with MEGA 7.0 software. Bootstrap values (1000 replicates) are shown as percentages at the branch nodes. The Arabic numerals I–IV represent different ARF groups. b Intron/exon structures of putative ARF genes in melon. Exons and introns are indicated by open boxes and lines, respectively. The results indicated that ARFs in melon could be divided into four groups: classes I, II, III, and IV
Tissue-specific expression patterns of CmARF genes
To investigate the transcript levels of these CmARFs in different tissues of melon plants, we collected samples of roots, leaves, stems, pistillate flower petals, fruits, and staminate flower petals. Expression analysis via semiquantitative PCR revealed ARF gene expression in those tissues, but the expression levels varied greatly among the different tissues (Fig. 5). CmARF19, CmARF3, and CmARF18-like1 were weakly expressed in the young roots; CmARF19-like, CmARF19, CmARF6, CmARF3, CmARF18-like1, CmARF16-like, CmARF8, CmARF2, and CmARF18-like3 were barely expressed in the stems; and CmARF19, CmARF3, and CmARF18-like1 were barely expressed in the leaves and female flowers. With the exception of CmARF18-like1, all the ARF genes were expressed in the fruits. The expression levels of CmARF2, CmARF16-like, CmARF18-like2, and CmARF19-like were especially high in the melon fruits, and CmARF6-like, CmARF5, CmARF9, CmARF17, CmARF1, CmARF4 and CmARF18 were expressed in all tissues. The expression levels of CmARF1, CmARF2, CmARF3, CmARF6, CmARF8, CmARF18-like3, and CmARF19-like were especially high in the pistillate flower petals.
Morphological changes in melon fruits during early development
In the absence of pollination, the fruits exhibited no significant change in either transverse diameter or longitudinal diameter throughout the entire process (Fig. 6a, b). The transverse and longitudinal diameters of the fruits gradually increased after growth regulator treatment and artificial pollination. On the third day of treatment, compared with those in the unpollinated group, the transverse and longitudinal diameters of the fruits in the two treatment groups increased rapidly, resulting in significant differences. During the entire treatment period, compared with those in the unpollinated group, the transverse and longitudinal diameters of the fruits in the growth regulator treatment group increased on average by 38.1% and 25.0%, respectively. Compared with those of the fruits resulting from unpollinated flowers, the average increases in the transverse and longitudinal diameters in the fruits after artificial pollination were 39.1% and 52.9%, respectively (Fig. 6a). The increase in the transverse diameters of the fruits treated with the growth regulator was highly significant, while the increase in the longitudinal diameters of the fruits resulting from artificially pollinated flowers was also highly significant. Images of the fruits during different periods of treatment also demonstrated these findings (Fig. 6c).
Growth changes on melon fruit after treatment with a growth regulator and artificial pollination. Early development of melon fruit. a Transverse diameters of melon fruits during early development. The results indicated that the transverse diameters of the fruits gradually increased after growth regulator treatment. b Longitudinal diameters of melon fruits during early development. The results indicated that the longitudinal diameters of the fruits gradually increased after artificial pollination. The data (means ± SDs) are the means of three replicates, with standard errors shown by vertical bars, n = 9. * and ** indicate significant differences at p < 0.05 and p < 0.01, respectively, between the unpollinated group and treatment groups according to Tukey’s test. c Shape change in melon fruits during the early development stage. The transverse and longitudinal diameters of the fruits gradually increased after growth regulator treatment and artificial pollination
CmARF expression during melon fruit development
We used qRT-PCR to analyze the expression of ARF genes during the early development of melon fruits after different treatments (Fig. 7). The results showed that, as fruit development progressed, the expression levels of CmARF1, CmARF4, and CmARF5 tended to increase in the three treatments, while those of CmARF2, CmARF9, and CmARF19 tended to decrease. The expression levels of CmARF3, CmARF6-like, CmARF8, CmARF16-like, CmARF17, CmARF18-like1, CmARF18-like2, and CmARF19-like tended to increase but then decrease. The trends of the other 3 ARF genes were not obvious.
Expression of ARF family genes in melon fruits after growth regulator and artificial pollination treatment. The mRNA expression levels of 17 ARF genes were determined by qRT-PCR. The figure shows the unpollinated group, the artificially pollinated group, and the growth regulator Fengchanji No. 2 treatment group. Fruits were collected at 0, 1, 2, 3, and 5 days after treatment. The data (means ± SDs) are the means of three replicates, with standard errors shown by vertical bars, n = 3. The different superscript letters imply a significant difference at p < 0.05 between the unpollinated group and the treatment group according to Tukey’s test. The results indicated that different CmARF genes exhibit different spatial and temporal expression patterns during the early fruit development of melon
The expression levels of CmARF9, CmARF16-like, and CmARF18-like2 in the two treatment groups were all greater than those in the unpollinated group at the same time points (Fig. 7). In addition, the expression of CmARF9 in the artificial pollination treatment was greater than that in the growth regulator treatment, and the expression of CmARF16-like exhibited the opposite trend. The expression level of CmARF5 in both treatment groups was greater than that in the unpollinated group. Moreover, the expression level of CmARF3 in both treatment groups was lower than that in the unpollinated group at first but then was greater than that in the unpollinated group at subsequent time points. The expression of CmARF1 and CmARF17 in the growth regulator treatment group was greater than that in the unpollinated group at all stages. The expression of CmARF1 in the artificial pollination group was first lower than that in the unpollinated group but then was greater than that in the unpollinated group. There was no obvious change in the expression levels of CmARF17 in the artificial pollination group. Moreover, the expression level of CmARF4 was lower than that in the unpollinated group beginning on the second day of growth regulator treatment, lower than that in the unpollinated group on the third day of artificial pollination treatment, and greater than that in the unpollinated group for the remainder of the time. The expression level of CmARF6 was lower than that in the unpollinated group on the 5th day after growth regulator treatment and greater than that in the unpollinated group for the remainder of the time, while the expression in the pollinated group was consistently greater than that in the unpollinated group. The expression level of CmARF6-like was lower than that in the unpollinated group on the third day after growth regulator treatment and greater for the remainder of the time. In addition, the expression level of CmARF6-like in the artificial pollination group was consistently greater than that in the unpollinated group beginning on the second day. The expression level of CmARF8 in both treatment groups was lower than that in the unpollinated group on the second day but was greater than that in the unpollinated group for the remainder of the time. Moreover, the expression level of CmARF18 in the growth regulator treatment group was lower than that in the unpollinated group on the 5th day but greater than that in the unpollinated group for the remainder of the time. The expression level of CmARF19-like in the growth regulator treatment group was greater than that in the unpollinated group on the 5th day but lower than that in the unpollinated group for the remainder of the time.
Correlation analysis between transverse or longitudinal diameter and variation of expression level of ARF members
Correlation analysis showed the coefficient between transverse diameter or longitudinal diameter and variation of expression level of CmARF9, CmARF19, CmARF2, and CmARF1 over 70% in the unpollination group and the two treatment groups (Tables 2 and 3). Moreover, the coefficient between transverse diameter and variation of expression level of CmARF16-like and CmARF3 was significant difference in correlation between unpollination group and the two treatment groups (Table 2), and the coefficient between longitudinal diameter and variation of expression level of CmARF5, CmARF16-like, and CmARF19-like was significant difference in correlation between unpollination group and the two treatment groups (Table 3). It suggested that CmARF9, CmARF16-like, CmARF19-like, CmARF19, CmARF1, CmARF2, CmARF3, and CmARF5 do great related to fruit growth.
Discussion
Auxin is associated with all stages of plant growth and development, such as cell division, cell elongation and differentiation, apical dominance, plant orientation, senescence, shedding, and, in certain plant species, the induction of blooming (Marc et al. 2007). Auxin regulates these processes by controlling gene expression via a series of uniquely functional DNA-binding ARFs (Mcphie 2002). In recent years, given the complete genome sequencing of multiple species, studies on the plant ARF family have been carried out gradually for different species. The identification of the ARF family began with Arabidopsis (Tamura et al. 2011), and the identification of ARF family members is the basis for studying the regulatory mechanism and function of ARFs in plant growth and development.
In this study, we identified 17 ARF genes in melon. There are fewer CmARF members in melon than in both Arabidopsis (23) and rice (25) (Tamura et al. 2011; Goff 2002). The genome size of melon is approximately the same as that of rice but is very different from that of Arabidopsis (melon: 450 Mb, Arabidopsis: 125 Mb, rice: 420 Mb) (Weng et al. 2017; Livak and Schmittgen 2001). Thus, the number of ARF genes in species with large genomes is not always large. Most members of the CmARF gene family have a complex structure with approximately 10 introns. Subcellular localization predicted that all of these genes except CmARF15 were localized in the nucleus. We also constructed a phylogenetic tree to analyze the relationships among ARF family genes in melon. In addition, CmARF8 and AtARF8 were highly homologous, reaching 96%. CmARF16-like, CmARF17, CmARF18, and CmARF18-like1 in melon contain a DBD and an auxin_resp domain but not a CTD (Aux/IAA-binding domain). Other CmARFs contain three typical domains: a DBD, an auxin_resp domain, and a CTD.
ARFs exhibit tissue-specific expression. In Arabidopsis, ARF9 is expressed in all tissues (Philip et al. 2004). In addition, GrARF1, GrARF2a, GrARF3a, GrARF4a, GrARF5a, GrARF6b, GrARF16c, GrARF17a, GrARF19.1a, and GrARF19.2 are highly expressed in flower buds of Gossypium raimondii, and GrARF1, GrARF4a, GrARF5a, GrARF16c, GrARF17a, and GrARF19.1a are highly expressed in the leaves (Sun et al. 2015). We performed semiquantitative PCR to analyze the differences in expression among different tissues during the early development of melon. We found that the expression levels of CmARF2, CmARF16-like, CmARF18-like2, and CmARF19-like were especially high in melon fruits. Moreover, the expression levels of CmARF1, CmARF2, CmARF3, CmARF6, CmARF8, CmARF18-like3, and CmARF19-like were especially high in pistillate flower petals; and CmARF1, CmARF4, CmARF5, CmARF6-like, CmARF9, CmARF17, and CmARF18 were expressed in all tissues. Thus, the expression of ARFs in melon is tissue specific. Studies have shown that ARF2 regulates the formation of floral organs (Finet et al. 2010) and that ARF5 is associated with embryonic development and vascular bundle formation (Weijers et al. 2006; Wenzel et al. 2010). The homologous genes of AtARF2 and AtARF5 in melon, CmARF2, and CmARF5, were identified. CmARF2 is highly expressed in fruits and female flowers, while CmARF5 is highly expressed in all parts. Therefore, we speculate that ARFs whose expression differs in different tissues have different functions during plant growth and development.
ARFs participate in the transcriptional regulation of fruit development (Johnson et al. 2007; Bohner and Bangerth 1988; De et al. 2008). In this study, the fruits resulting from the pollinated flowers quickly expanded, while the unpollinated ovaries did not grow. CmARF9, CmARF16-like, CmARF19-like, CmARF19, CmARF1, CmARF2, CmARF3, and CmARF5 may be associated with the fruit growth of melon during early development. In tomato, ARF9 negatively controls cell division during early fruit development. The transcription level of SlARF9 increased within 48 h after pollination but subsequently decreased in the following days (Wang et al. 2005). The high expression of CmARF9 before pollination decreased after artificial pollination or growth regulator treatment in this study, similar to that of SlARF9 in tomato. ARF19 is induced by either IAA or ethylene treatments, not only participates in auxin signaling, but also plays a critical role in ethylene responses in Arabidopsis (Jisheng Li et al. 2019). Decreased expression of CmARF9 may be related to cell division and early fruit development. It is worth noting that in the unpollinated ovaries, ARF9 expression decreased sharply 1 day after flowering, which was significantly faster than that in the treatment group. We hypothesized that ARF9, in addition to promoting the regulation of cell division to regulate fruit enlargement, was also involved in other biochemical reactions in plants. ARF16 has been shown to be involved in the regulation of root crown cell differentiation in Arabidopsis (Wang et al. 2005). ARF3 regulates floral meristem determinacy by repressing cytokinin biosynthesis and signaling (Zhang et al. 2018). ARF3 also regulates compound leaf patterns in Medicago truncatula (Peng et al. 2017), and ARF5 has been shown to mediate root meristem formation in Arabidopsis and poplar (Berleth and Jurgens 1993; Johnson and Douglas 2007). However, relationships between ARF16-like, ARF3, and ARF5 and fruit development have not been reported, which warrants further discussion in the future. It is interesting that the transverse diameter of the fruit obviously increased after growth regulator treatment, while the longitudinal diameter of the fruit obviously increased after artificial pollination. By analyzing the expression of ARFs during fruit development of melon, we found that the expression of CmARF9 in the artificial pollination treatment was greater than that in the growth regulator treatment, and CmARF16-like exhibited the opposite trend. The expression of CmARF3 in the growth regulator treatment was greater than that in the artificial pollination treatment after 2 days, and the opposite trend was observed on day 5 after treatment. Based on the changes in ARF expression and fruit growth in each group, it was suggested that CmARF16-like and CmARF3 were correlated with the horizontal cell growth of melon fruit, while CmARF9 was correlated with the vertical cell growth of melon fruit. The duration, rate, and direction of cell division in developing ovaries may have significantly affected the final fruit size and shape (Bohner and Bangerth 1988). Hence, the relationship of the mechanism between ARFs and the transverse or longitudinal diameter needs further study.
SlARF8 and SlARF7 in tomato are involved in auxin signal transduction during fruit set and development (De et al. 2008). ARF8 transcription in Arabidopsis thaliana and tomato increases during floral development, and the expression level remains high in unpollinated ovaries but decreases after pollination (Marc et al. 2007). In this study, the expression level of CmARF8 in the pollinated group and the growth regulator treatment group was lower than that in the unpollinated group on the second day after pollination but greater than that in the unpollinated group during the rest of the time. Before pollination, the level of CmARF8 expression was low, but it increased after pollination or after growth regulator treatment. These findings contrast with results in tomato (Marc et al. 2007), possibly because ARF8 functions differently in different species.
Conclusion
In summary, we proposed a model to explain this phenomenon. When the ovary was not pollinated, Aux/IAA suppressors formed homologous dimers with ARFs; consequently, ARF activity was inhibited, and auxin signal transduction was inhibited, resulting in fruit failure. After ovary pollination, auxin increases rapidly, Aux/IAA inhibitors are degraded by the ubiquitination pathway, ARFs are released, auxin response genes are activated, and then fruit set or fruit development is initiated. Verification of this prediction and investigation of other possible modes of action of ARFs will constitute our future research goals.
Abbreviations
- ARF:
-
Auxin response factor
- Aux/IAA:
-
Auxin/indole-3-acetic acid
- GH3:
-
Gretchen Hagen 3
- SAUR:
-
Small auxin-up RNA
- AuxRE-core:
-
Core element of the auxin response region
- AuxRE:
-
Auxin response element
- DBD:
-
DNA-Binding domain
- CTD:
-
C-Terminal dimerization domain
- MR:
-
Middle region
- AD:
-
Activation dimer
- RD:
-
Repression dimer
- qRT-PCR:
-
Quantitative real-time PCR
References
Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37:W202–W208. https://doi.org/10.1093/nar/gkp335
Berleth T, Jurgens G (1993) The role of the monopteros gene in organising the basalbody region of the Arabidopsis embryo. Development 118:575–587. https://doi.org/10.1016/0168-9525(93)90246-E
Bohner J, Bangerth F (1988) Effects of fruit set sequence and defoliation on cell number, cell size and hormone levels of tomato fruits (Lycopersicon esculentummill.) within a truss. Plant Growth Regul 7:141–155. https://doi.org/10.1007/bf00028237
Davies PJ (1987) The plant hormones: their nature, occurrence, and functions. Plant Horm Physiol Biochem Mol Biol:1–2. https://doi.org/10.1007/978-94-011-0473-9_1
De JM, Wolters-Arts MR, Mariani C, Vriezen WH (2008) The solanum lycopersicum auxin response factor 7 (SlARF7) regulates auxin signaling during tomato fruit set and development. Plant J57:160–170. https://doi.org/10.1111/j.1365-313X.2008.03671.x
Dolf W, Eva B, Ger KE, Alexandra S, Thorsten H, Marika K, Wilmoth JC, Reed JW, Gerd J (2014) Developmental specificity of auxin response by pairs of arf and aux/iaa transcriptional regulators. EMBO J 24:1874–1885. https://doi.org/10.1038/sj.emboj.7600659
Ellis CM, Punita N, Young JC, Gretchen H, Guilfoyle TJ, Reed JW (2005) Auxinresponsefactor1 and auxinresponsefactor2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development:132, 4563–4574. https://doi.org/10.1242/dev.02012
ElsharkawyI SSM, Jones B, Mila I, Kumar PP, Bouzayen M, Jayasankar S (2014) Tir1-like auxin-receptors are involved in the regulation of plum fruit development. J Exp Bot 65:5205–5215. https://doi.org/10.1093/jxb/eru279
Finet C, Fourquin C, Vinauger M, Berne-Dedieu A, Chambrier P, Paindavoine S, Scutt CP (2010) Parallel structural evolution of auxin response factors in the Tngiosperms. Plant J63:952–959. https://doi.org/10.1111/j.1365-313x.2010.04292.x
Garcia-Mas J, Benjak A, Sanseverino W, Bourgeois M, Mir G, González VM, Hénaff E, Câmara F, Cozzuto L, Lowy E, Alioto T, Capella-Gutiérrez S, Blanca J, Cañizares J, Ziarsolo P, Gonzalez-Ibeas D, Rodríguez-Moreno L, Droege M, Du L, Alvarez-Tejado M, Lorente-Galdos B, Melé M, Yang L, Weng Y, Navarro A, Marques-Bonet T, Aranda MA, Nuez F, Picó B, Gabaldón T, Roma G, Guigó R, Casacuberta JM, Arús P, Puigdomènech P (2012) The genome of melon (Cucumis melo L.). Proc Natl Acad Sci 109:11872–11877. https://doi.org/10.1073/pnas.1205415109
Goetz M, Hooper LC, Johnson SD, Rodrigues JCM, Viviansmith A, Koltunow AM (2007) Expression of aberrant forms of stimulates parthenocarpy in Arabidopsis and tomato. Plant Physiol 145:351–366. https://doi.org/10.1104/pp.107.104174
Goff SA (2002) A draft sequence of the rice genome (Oryza sativaL. ssp. japonica). Science 296:92–100. https://doi.org/10.1126/science.1068275
Guilfoyle TJ, Hagen G (2007) Auxin response factors. Curr Opin Plant Biol 10:453–460. https://doi.org/10.1007/s003440010026
Guo AY, Zhu QH, Xin C (2007) Gsds:a gene structure display server. Hereditas 29:1023–1026. https://doi.org/10.1360/yc-007-1023
Li J, Dai X, Zhao Y (2019) A role for auxin response factor 19 in auxin and ethylene signaling in Arabidopsis. Plant Physiol 140:899–908 http://www.plantphysiol.org/content/plantphysiol/140/3/899
Johnson LA, Douglas CJ (2007) Populus trichocarpa monopteros/auxinresoonsefactor5 (ARF5) genes: comparative structure, sub-functionalization, and populus-arabidopsis microsynteny. Can J Bot 85:1058–1070. https://doi.org/10.1139/B07-065
Lim PO, Lee IC, Kim J, Kim HJ, Ryu JS, Woo HR, Hong GN (2010) Auxin response factor 2 (ARF2) plays a major role in regulating auxin-mediated leaf longevity. J Exp Bot 61:1419–1430. https://doi.org/10.1093/jxb/erq010
Liu Y, Jiang HY, Chen WJ, Qian YX, Ma Q, Cheng BJ, Zhu SW (2011) Genome-wide analysis of the auxin response factor (ARF) gene family in maize (Zea mays). Plant Growth Regul 63:225–234. https://doi.org/10.1007/s10725-010-9519-0
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 4:402–408. https://doi.org/10.1006/meth.2001.1262
Marc G, Adam VS, Johnson SD, Koltunow AM (2006) Auxinresponsefactor8 is a negative regulator of fruit initiation in Arabidopsis. Plant Cell 18:1873–1886. https://doi.org/10.1105/tpc.105.037192
Marc G, Hooper LC, Johnson SD, Rodrigues JCM, Adam VS, Koltunow AM (2007) Expression of aberrant forms of auxinresponsefactor8 stimulates parthenocarpy in Arabidopsis and tomato. Plant Physiol 145:351–366. https://doi.org/10.1104/pp.107.104174
Mcphie P (2002) The protein protocols handbook. Anal Biochem 315:289–289. https://doi.org/10.1016/S0003-2697(02)00682-6
Park JE, Park JY, Kim YS, Staswick PE, Jeon J, Yun J, Kim SY, Kim J, Lee YH, Park CM (2007) Gh3-mediated auxin homeostasis links growth regulation with stress adaptation response in Arabidopsis. J Biol Chem 282:10036–10046. https://doi.org/10.1074/jbc.M610524200
Peng J, Berbel A, Madueño F, Chen R (2017) Auxinresponsefactor3 regulates compound leaf patterning by directly repressing palmate-likepentafoliat1 expression in Tedicago truncatula. Front Plant Sci, 8:1630. https://doi.org/10.3389/fpls.2017.01630
Philip Z, Matthias HH, Lars H, Wilhelm G (2004) Genevestigator. Arabidopsis microarray database and analysis toolbox. Plant Physiol 136:2621–2632. https://doi.org/10.1104/pp.104.046367
Piya S, Shrestha SK, Binder B, Stewart CN Jr, Hewezi T (2014) Protein-protein interaction and gene co-expression maps of ARFs and aux/IAAs in Arabidopsis. Front Plant Sci 5. https://doi.org/10.3389/fpls.2014.00744
Shen C, Yue R, Sun T, Zhang L, Cu LQ, Tie SG, Wang HZ, Yang YJ (2015) Genome-wide identification and expression analysis of auxin response factor gene family in Medicago truncatula. Front Plant Sci 6:1–13. https://doi.org/10.3389/fpls.2015.00073
Sun R, Wang K, Guo T, Jones DC, Cobb J, Zhang B, Wang Q (2015) Genome-wide identification of auxin response factor (ARF) genes and its tissue-specific prominent expression in Tossypium raimondii. Funct Integr Genomics 15:481–493. https://doi.org/10.1007/s10142-015-0437-0
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) Mega5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mo Biol Evol 28:2731-2739.https://doi.org/10.1093/molbev/msr121
Tatematsu K, Kumagai S, Muto H, Sato A, Watahiki MK, Harper RM, LiscumE YKT (2004) Massugu2 encodes aux/iaa19, an auxin-regulated protein that functions together with the transcriptional activator nph4/arf7 to regulate differential growth responses of hypocotyl and formation of lateral roots in Arabidopsis thaliana. Plant Cell 16:379–393. https://doi.org/10.1105/tpc.018630
Thorsten H, Eva B, Isabel BU, Marika K, Gerd J (2002) The Arabidopsis bodenlos gene encodes an auxin response protein inhibiting monopteros-mediated embryo patterning. Genes Dev 16:1610–1615. https://doi.org/10.1101/gad.229402
Tiwari SB, Gretchen H, Tom G (2003) The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15:533–543. https://doi.org/10.1105/tpc.008417
Tiwari SB, Wang XJ, Hagen G, Guilfoyle TJ (2001) Aux/iaaproteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell 13:2809–2822. https://doi.org/10.1105/tpc.010289
Ulmasov T, Hagen G, Guilfoyle TJ (1999) Activation and repression of transcription by auxin-response factors. Proc Natl Acad Sci U S A 96:5844–5849. https://doi.org/10.1016/B978-012373983-4.50013-4
Vanneste S, Friml J (2009) Auxin: atrigger for change in plant development. Cell 136:1005–1016. https://doi.org/10.1016/j.cell.2009.03.001
Wang JW, Wang LJ, Mao YB, Cai WJ, Xue HW, Chen XY (2005) Control of root cap formation by microRNA-targeted auxin response factors in Arabidopsis. Plant Cell 17:2204–2216. https://doi.org/10.1105/tpc.105.033076
Weijers D, Schlereth A, Ehrismann JS, Schwank G, Kientz M, Jürgens G (2006) Auxin triggers transient local signaling for cell specification in Arabidopsis embryogenesis. Dev Cell 10:265–270. https://doi.org/10.1016/j.devcel.2005.12.001
Weng Q, Song J, Ma H, Yuan J, Liu Y (2017) Cloning and expression analysis of zmabi3 gene in Zea mays. Turk J Biochen 42. https://doi.org/10.1515/tjb-2016-0303
Wenzel CL, Mathias S, Qian Y, Jim M (2010) Dynamics of monopteros pteros and pin-rormed1 expression during leaf vein pattern formation in Arabidopsis thaliana. Plant J 49:387–398. https://doi.org/10.1111/j.1365-313X.2006.02977.x
Wilmoth JC, Shucai W, Tiwari SB, Joshi AD, Gretchen H, Guilfoyle TJ, Alonso JM, Ecker JR, Reed JW (2010) Nph4/arf7 and arf19 promote leaf expansion and auxin-induced lateral root formation. Plant J Cell Mol Biol 43:118–130. https://doi.org/10.1111/j.1365-313X.2005.02432.x
Yang C, Xu M, Xuan L, Jiang X, Huang M (2014) Identification and expression analysis of twenty ARF genes in Populus[J]. Gene 544:134–144. https://doi.org/10.1016/j.gene.2014.04.067
Yoko O, Hidehiro F, Makoto O, Athanasios T, Masao T (2007) arf7 and arf19 regulate lateral root formation via direct activation of lbd/asl genes in Arabidopsis. Plant Cell 19:118–130. https://doi.org/10.1105/tpc.106.047761
Zhang K, Wang R, Zi H, Li Y, Cao X, Li D, Guo L, Tong J, Pan Y, Jiao Y (2018) Auxinrespovsefactor3 regulates floral meristem determinacy by repressing cytokinin biosynthesis and signaling. Plant Cell 30. https://doi.org/10.1105/tpc.17.00705
Zhang S, Chen S, Chen F, Fang W (2012) The regulatory role of the auxin in the creeping chrysanthemum habit. Russ J Plant Physiol 59:364–371. https://doi.org/10.1134/S1021443712020203
Funding
This work was financially supported by the Young Scientists Fund of the National Natural Science Foundation of China (grant No. 31201641), the 2018 Joint Funding Project of the Beijing Natural Science Foundation-Municipal Education Commission (KZ201810020027), the Science and Technology Committee of Beijing (Z131105000413), and the Earmarked Fund for Beijing Leaf Vegetables Innovation Team of Modern Agro-industry Technology Research System (BAIC08).
Author information
Authors and Affiliations
Contributions
Bei Wu, Lu Wang, Gaoyang Pan, and Jinghong Hao conceived and designed the experiments. Bei Wu, Lu Wang, and Gaoyang Pan performed the experiments. Ting Li analyzed the data and prepared some figures and tables. Xin Li provided advice on the analysis and interpretation of the data. Lu Wang and Gaoyang Pan prepared some figures and tables. Bei Wu, Lu Wang, and Gaoyang Pan wrote the paper. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Handling Editor: Liwen Jiang
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wu, B., Wang, L., Pan, G. et al. Genome-wide characterization and expression analysis of the auxin response factor (ARF) gene family during melon (Cucumis melo L.) fruit development. Protoplasma 257, 979–992 (2020). https://doi.org/10.1007/s00709-020-01484-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00709-020-01484-2