Abstract
All organisms are able to modulate gene expression in response to internal and external stimuli. Trypanosomes represent a group that diverged early during the radiation of eukaryotes and do not utilise regulated initiation of transcription by RNA polymerase II. Here, the mechanisms present in trypanosomes to alter gene expression in response to stress and change of host environment are discussed and contrasted with those operating in yeast and cultured mammalian cells.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In eukaryotes, gene expression changes in response to alterations to the environment. The alteration can result from both extrinsic signals, for example, a change in nutrients, and intrinsic changes, such as a timing mechanism. The immensely complex temporal and spatial patterns of gene expression integral to the development of multicellular organisms occur in response to extrinsic and intrinsic signals that have evolved to produce the spatial arrangement of the different cell types that comprise the organism. Unicellular organisms are clearly simpler but are also able to change gene expression in two key ways: first, to optimise the survival of one cell type in response to changes in the environment and, second, to trigger a pathway leading to a different developmental form in response to other signals. Alterations in gene expression can be achieved by a change in one or more of the following: transcription, usually an alteration in the frequency of initiation; the fraction of pre-mRNA that undergoes successful maturation and export from the nucleus; cytoplasmic mRNA half-life; and the fraction of mRNA in translation and the efficiency of that translation, the latter being usually determined by the frequency of initiation. In addition, the activity of the protein product can be modulated.
Yeasts are the best-characterised unicellular eukaryotes and, despite being secondarily evolved from hyphal fungi, are good exemplars for the characteristics typical of a unicellular organism. A growing budding yeast cell contains one or two copies of each of the ∼6,000 genes, around 30,000 mRNA and 5 × 107 protein molecules (10 pg cell, 50% protein) (http://bionumbers.hms.harvard.edu/). So, a single gene encoding a moderately abundant protein (0.1% total protein) at steady-state levels is responsible for 30 mRNA and 50,000 protein molecules. Changes in the rate of transcription, and thus the number of mRNA molecules, will have a profound effect on the production of a protein as each mRNA is translated many times. Most yeast mRNAs have half-lives of tens of minutes and a transcriptional response must therefore operate on a similar timescale. Any rapid response mechanism has to take account of the mRNA already in the cytoplasm and such responses must be post-transcriptional, at least in the initial phase.
In animal cells, a similar combination of transcriptional and post-transcriptional responses occurs. However, the transcriptional response is often more complex, involving a greater number of genes, as it has evolved to optimise the survival of the organism. In contrast, the absence of the appropriate response to stress in a unicellular organism results in death of the whole organism.
Members of the Kinetoplastida, such as trypanosomes, are amongst the best-characterised protozoa. Initial investigations aimed to determine the molecular mechanisms of virulence. Subsequently, more molecular details of their basic cell biology have emerged along with the realisation that kinetoplastids are an excellent model of divergent eukaryotes as they are as evolutionarily distant from animals, plants and fungi as any other eukaryote (Dacks et al. 2008). One feature of the basic biology that reflects this evolutionary divergence is the mechanism of gene expression and the downstream consequences for regulation. In mammals and yeast, transcription of protein-coding genes is monocistronic and performed by RNA polymerase (RNAP) II, while in nematodes RNAP II transcription of protein-coding genes is monocistronic or polycistronic, with up to six genes transcribed from a single promoter. In kinetoplastid protozoa, polycistronic transcription predominates: most protein-coding genes are present in transcription units of tens or sometimes hundreds of genes (Martinez-Calvillo et al. 2003; Martinez-Calvillo et al. 2004; Siegel et al. 2009; Kolev et al. 2010). The pre-mRNA is co-transcriptionally processed to monocistronic mRNAs through coupled steps that trans-splice a 39-nucleotide exon to the 5′ end of the mRNA and cleave and polyadenylate the 3′ end of the upstream mRNA (LeBowitz et al. 1993; Ullu et al. 1993; Matthews et al. 1994). The spliced leader RNA (SL RNA) containing the 39-nucleotide exon is transcribed by RNAP II (Gilinger and Bellofatto 2001) and is capped (Bangs et al. 1992); thus, trans-splicing adds the 5′ cap to the mRNA, and the mature mRNA is therefore structurally similar to other eukaryotic mRNAs. Estimates of the number of mRNA and protein molecules are similar to yeast and most mRNAs have half-lives of tens of minutes (Haanstra et al. 2008).
Despite >99% of protein-coding genes being transcribed by RNAP II, their promoters are uncharacterised, although likely to be located at the 5′ end of gene arrays and probably marked epigenetically as opposed to by sequence (Siegel et al. 2009). In most cases, genes acting in a particular pathway are randomly distributed throughout the genome; so, it is unlikely that operon-like transcriptional control occurs. Importantly, it is believed that nearly all protein-coding genes are transcribed by RNAP II at a similar rate and high RNA synthesis rates are obtained either through amplification of gene copy number, for example, 19 copies each of the α- and β-tubulin genes (Ersfeld et al. 1998), or, in T. brucei, transcription of a small number of protein-coding genes by the highly processive RNAP I, most notably those genes encoding the mRNAs for the super-abundant cell surface proteins VSG and procyclin (Gunzl et al. 2003). Trans-splicing negates a requirement for RNAP II transcription to add a 5′ cap.
Absence of RNAP II regulation implies that the final level of any protein must be determined by one or more post-transcriptional mechanisms (reviewed in Clayton and Shapira 2007). The sequence around the trans-splice acceptor site can alter the splicing efficiency and consequently the levels of an mRNA (Siegel et al. 2005) and alternative splicing occurs (Nilsson et al. 2010), but nothing is known about how this may be modulated in response to a changing environment. Variation in mRNA half-lives underpin some of the differential gene expression in the various developmental forms (see, for example, Webb et al. 2005). There are some clear examples of translational regulation (Walrad et al. 2009). However, there is no clearly elucidated pathway linking a change in gene expression to an extrinsic or intrinsic signal.
Here, the response to three different extrinsic signals in trypanosomes is discussed. The first two, heat shock and the unfolded protein response (UPR), were almost certainly present in the last common eukaryotic ancestor and the third, differentiation from one life cycle stage to the next, has probably evolved since divergence but is paralogous to similar transitions on most other eukaryotes. In animals and yeasts, each of these responses contains a substantial transcriptional element and how the trypanosome might respond in the absence of transcriptional control is discussed. There are little hard data for trypanosomes in this area and much of the discussion concerning the possibilities in trypanosomes below should be treated as speculations, hopefully well informed.
The heat shock response in eukaryotes with heat-inducible transcriptional control
Heat shock causes denaturation and aggregation of proteins leading to perturbation of many cellular processes. It disrupts the Golgi complex and changes the morphology of mitochondria, nucleoli and the cytoskeleton (Welch and Suhan 1985). To overcome the damage, chaperone proteins that protect against oxidative damage and protective heat shock proteins (HSPs) are preferentially synthesised. Transcription and translation of housekeeping genes is shut down in order to conserve anabolic energy for the repair processes.
Central to the heat shock response of many eukaryotes are heat shock transcription factors (HSF) that bind to a conserved region in heat-inducible promoters called the heat shock element. Saccharomyces cerevisiae (Sorger and Pelham 1988) and Drosophila (Clos et al. 1990) express one, mammalian cells three (Pirkkala et al. 2001) and plants more than 20 heat shock transcription factors (von Koskull-Doring et al. 2007). In unstressed mammalian cells, the master regulator of heat-inducible gene expression, HSF1, is in a complex with HSP90. Upon heat shock, HSP90 associates with the increasing number of denatured proteins releasing HSF1 (Zou et al. 1998) that trimerises and translocates from the cytoplasm to the nucleus, leading to the transcription of genes coding for heat shock proteins (Sarge et al. 1993). Phosphorylation is central to the regulation of HSF1 activity. On one hand, phosphorylation of HSF1 is required for translocation to the nucleus (Kim et al. 2005) and promotes transcriptional activity (Holmberg et al. 2001; Guettouche et al. 2005) but, on the other hand, phosphorylation at other sites can repress activity (Knauf et al. 1996; Kline and Morimoto 1997). Upon heat shock, HSF1 undergoes sumoylation mediated by a phosphorylation-dependent sumoylation motif (Hietakangas et al. 2006) but it is still disputed if sumoylation is an activating (Hong et al. 2001) or repressing modification (Hietakangas et al. 2006). The transcriptional activity of HSF1 bound to the heat shock element is repressed by HSP70 binding (Shi et al. 1998a); whether the binding of HSP70 prevents full activation of HSF1 or is involved in shutting off the transcriptional heat shock response is unknown. The transcriptional response cannot serve as an immediate response for the reasons discussed above.
For the immediate response, heat shock induces drastic changes in mRNA metabolism. mRNA splicing inhibition was observed in S. cerevisiae (Yost and Lindquist 1991) and human cell lines (Shin et al. 2004). In mammalian cells, the mechanism of inhibition is through the splicing factor SRp38 which is dephosphorylated and consequently interacts with the U1 small nuclear ribonucleoprotein particle interfering with the 5′-splice-site recognition (Shin et al. 2004). Heat shock also leads to nuclear accumulation of polyadenylated mRNA in both yeast (Saavedra et al. 1996) and mammalian cells (Gallouzi et al. 2000). Despite the general inhibition of mRNA splicing, mRNAs encoding HSPs are still processed and exported during heat shock. Some other changes are due to altered mRNA stability. In HeLa cells, HSP70 mRNA has a half-life of 50 min; it is relatively unstable for a cultured mammalian cell but increases at least tenfold on heat shock (Theodorakis and Morimoto 1987). HSP70 protein also binds AUF1, an mRNA-binding protein that destabilises some mRNAs containing the regulatory ARE element, and sequesters AUF1 in the nucleus, resulting in the stabilisation of the mRNA targets of AUF1 (Laroia et al. 1999).
Heat shock causes the formation of stress granules in the cytoplasm containing mRNA bound to stalled ribosomal pre-initiation complexes and, depending on cell type and organism, a variety of RNA-binding proteins, RNA helicases, mRNA degradation enzymes and kinases. The large ribosomal subunits are not found in stress granules (Anderson and Kedersha 2009). Sequestration of mRNAs to stress granules may contribute to general translational repression and store mRNAs for heat shock recovery. It is assumed that the heat shock stress granules are sorting places where mRNAs are either sent to be degraded and/or to be stored. Not all mRNAs localise to stress granules upon heat shock: in human cells, the HSP70 and HSP90 mRNA were demonstrated to be excluded from stress granules (Kedersha and Anderson 2002; Stohr et al. 2006).
The stress granules that form in response to different types of environmental stress are not the same and the repression of translation common to most stresses occurs through different pathways. The best-understood mechanism is dependent on the phosphorylation of eIF2α (McEwen et al. 2005; Kedersha et al. 1999). Phosphorylated eIF2α forms an inactive complex with the guanine nucleotide exchange factor eIF2B so that recycling of the GDP-bound eIF2α into the translationally active GTP-bound form cannot occur and cap-dependent translation initiation is diminished. In mammalian cells, stress granule formation on heat shock is dependent on eIF2α phosphorylation (Kedersha et al. 1999; Farny et al. 2009), whereas in Drosophila, budding yeast and trypanosomes it is not and occurs through a pathway that has yet to be identified (Kramer et al. 2008; Farny et al. 2009; Grousl et al. 2009). Selective escape of individual mRNAs from the heat-shock-induced repression of translation can occur through cap-independent initiation; the 5′-UTR of HSP70 mRNA contains an internal ribosome entry site and the mRNA is translated during heat shock (Rubtsova et al. 2003).
The heat shock response in kinetoplastids
Changes in temperature occur regularly in the life cycle of trypanosomes, both in the arthropod vector and when the parasite is transmitted between the vector and mammalian host. Temperature shift is one of the factors triggering differentiation in trypanosomes and is discussed below. However, heat shock, where the cell ceases to proliferate and increases the expression of genes linked to recovery and survival, is more likely to occur in the poikilothermic vector, for example a tsetse fly in the full sun. The change in gene expression in response to heat shock occurs within a few minutes and provides an amenable system to investigate a change in gene expression in response to an external environmental change.
In Trypanosoma brucei, HSP70 mRNAs accumulate upon heat shock but this accumulation is clearly not due to transcriptional control (Lee 1995). Heat shock inhibits trans-splicing of the tubulin transcript (Muhich and Boothroyd 1988), whereas trans-splicing of the HSP70 mRNA is unaffected (Muhich and Boothroyd 1989). This difference is reflected in the HSP70 and tubulin mRNA levels over a time course of heat shock: HSP70 does not decrease whereas tubulin does (Kramer et al. 2008). The mechanism of the selective regulation of trans-splicing is not understood. Only the genes for a putative RNA helicase and a poly(A) polymerase contain a cis-intron (Mair et al. 2000; Berriman et al. 2005; Jae et al. 2010). Regulation of cis-splicing of the poly(A) polymerase on heat shock could potentially lead to a general inhibition of mRNA maturation as trans-splicing and polyadenylation are coupled processes, but it is unknown if there is any regulation. However, trans-splicing of the poly(A) polymerase gene gives two products: (a) the mature fully spliced mRNA and (b) a trans-spliced mRNA containing just the second exon (Siegel et al. 2010). It has not been tested if the latter mRNA is translated and if the truncated poly(A) polymerase can inhibit the activity of the polyadenylation process under some circumstances.
Heat shock also causes a rapid cessation of proliferation that coincides with a reduction in the steady-state levels of most mRNAs; so, by 2 h of heat shock, there is a 75% decrease in cellular mRNA resulting from decreased production and increased decay (Muhich and Boothroyd 1988; Kramer et al. 2008). In contrast, the steady-state levels of selected mRNAs, including HSP83 and HSP70, are maintained or increased (Lee 1998; Kramer et al. 2008). The 3′ UTR is required to maintain HSP70 mRNA levels at heat shock conditions (Hausler and Clayton 1996; Lee 1998). In two Leishmania species, the 3′ UTR of the HSP83 mRNA is sufficient for increased stability and translation on heat shock, whereas the 5′ UTR has no effect by itself but does act synergistically with the 3′ UTR (Zilka et al. 2001; Larreta et al. 2004). In Leishmania amazonensis, a cis-element sufficient for preferential translation upon heat shock was found in the 3′ UTR of the HSP83 mRNA. Mutagenesis based on the predicted structure abolished thermal regulation; it was suggested that the temperature-induced changes in the structure of the element might directly regulate the translation (David et al. 2009).
Heat shock also causes a decrease in polysomes that is probably necessary for the reduction in total mRNA and causes substantial changes in cytoplasmic ribonucleoprotein granules (Kramer et al. 2008). Processing (P)-bodies contain enzymes of the 5′–3′ mRNA degradation pathway and are increased in response to heat shock, probably resulting from the release of mRNA from polysomes. In addition, stress granules containing many of the proteins involved in the initiation of translation appear. XRNA is an abundant cytoplasmic 5′–3′ exoribonuclease capable of degrading mRNAs and is usually concentrated in P-bodies. However, on heat shock, it also forms a focus located at the posterior pole of the cell of unknown function (Kramer et al. 2008). Stress granule formation upon heat shock is independent of eIF2α phosphorylation (Kramer et al. 2008). In yeast and humans, some stresses induce tRNA cleavage (Thompson and Parker 2009) and the cleavage product induces stress granule assembly independent of eIF2α phosphorylation (Emara et al. 2010). Whether a similar mechanism exists in trypanosomes has not been tested although tRNA-derived small RNAs are present in cytoplasmic granules in Trypanosoma cruzi (Garcia-Silva et al. 2010).
In summary, the trypanosome response to heat shock is similar to other eukaryotes in that it is composed of a selective increase in the expression of a small number of genes in a background of generally decreased expression of the vast majority of other genes. In the absence of transcriptional regulation, the up-regulated genes have to avoid the general inhibition of pre-mRNA processing, the increased mRNA turnover and suppression of translation. The immediate response is not very different from other eukaryotes. The transcriptional response present in yeast and mammalian cells has no equivalent in trypanosomes except that it may have been replaced with selective mRNA stabilisation via post-transcriptional mechanisms. It would be interesting to determine whether trypanosomes can tolerate heat shock for a shorter time than yeast.
In trypanosomes, the recovery from heat shock takes several hours before growth resumes (Kramer et al. 2008). During the recovery period, cellular mRNA levels return to levels before heat shock. How the trypanosome monitors the level of mRNA is unknown, but feedback mechanisms have been reported in other eukaryotes. In yeast, Dcp1 is a subunit of the decapping enzyme that catalyses the first step in 5′–3′ mRNA decay and inactivation drastically slows the degradation of many mRNAs but does not alter steady-state levels (Muhlrad and Parker 1999). A similar experiment provides evidence for a feedback mechanism monitoring total mRNA in trypanosomes; depletion of CAF1, the catalytic subunit of the major deadenylase, strongly inhibits mRNA degradation of housekeeping and developmentally regulated mRNAs but has no effect on steady-state levels (Schwede et al. 2008; Schwede et al. 2009).
In yeast, there is some evidence that the feedback pathway may operate through Rpb4 and Rpb7, two non-core components of the RNAP II that shuttle between nucleus and cytoplasm (Selitrennik et al. 2006). Rpb4 and Rpb7 have a role in the initiation of transcription (Edwards et al. 1991; Orlicky et al. 2001) and 3′-end processing of mRNAs (Runner et al. 2008). Loading of Rpb4 and Rpb7 onto the mRNA requires interaction with the RNAP II core (Goler-Baron et al. 2008). Phosphorylation-dependent ubiquitinylation of two core components of RNAP II ejects the Rpb4/7 heterodimer from the polymerase core (Daulny et al. 2008). It is not known if this is a regulatory mechanism for repressing transcription or a way to release Rpb4/7 from the polymerase core. Rpb4/7 is exported with the transcript. In the cytoplasm, Rpb4/7 is required for efficient translation (Harel-Sharvit et al. 2010). In addition, Rpb4/7 has several roles in mRNA degradation: it stimulates deadenylation, recruits activators of decapping and is required for efficient degradation by the 3′–5′ degradation pathway and the 5′–3′ degradation pathway (Lotan et al. 2005; Lotan et al. 2007). Rpb4 und Rbp7 are also P-body components (Lotan et al. 2005; Lotan et al. 2007).
Investigation of cis-elements in mRNAs has not yet identified discrete motifs but has revealed regulation at both mRNA stability and translation. In Leishmania, the HSP70 locus contains six genes that are all transcribed at similar rates (Quijada et al. 1997). Five are HSP70-Ι genes and mRNA abundance increases after heat shock; the 3′ UTR is sufficient for the increase (Quijada et al. 2000). A fraction of HSP70-Ι mRNAs is bound to polysomes at both normal and heat shock temperatures (Folgueira et al. 2005). In contrast, the HSP70-ΙΙ mRNA has a different 3′ UTR and is not translated at normal temperatures. On heat shock, the mRNA abundance does not change but the mRNA is now translated (Folgueira et al. 2005). A chloramphenicol acetyl transferase (CAT) reporter with HSP70-ΙΙ 3 ′ UTR does not show any translational regulation upon temperature change (Folgueira et al. 2005). However, it was subsequently found that mRNAs with a neomycin phosphotransferase open reading frame and HSP70-ΙΙ 3′-UTR obtained by targeted replacement of the endogenous HSP70-ΙΙ gene display the same temperature-dependent translational regulation as endogenous HSP70-ΙΙ mRNA (Folgueira and Requena 2007); the former is a good example of misleading results that sometimes occur when reporters are used. Many changes in the transcriptome due to heat shock, oxidative stress, unfolded protein response and hyperosmotic shock are transient (Gasch et al. 2000); the large, transient changes in transcript levels at the beginning of a stress response may be required to set the protein levels to a new steady state. After reaching the new steady-state protein profile, high transcript levels are not required anymore to maintain the steady-state protein levels and the transcript levels can be reduced. Thus, a time course analysis is essential to characterise any stress response; this is not always included in studies.
Little is known about the molecular detail of the switch that triggers the heat shock response in trypanosomes. However, one of the initial events of heat shock is conserved; one HSP70 isoform translocates from the cytoplasm to the nucleus in T. cruzi (Martin et al. 1993; Olson et al., 1994). The response includes dramatic changes in cytoplasmic mRNP granules but the precise function of P-bodies and stress granules remains unclear. It can be speculated that concentrating mRNA degradation in specific foci like P-bodies may be important for the selective degradation of different mRNAs and some mRNAs may be stored in stress granules during the heat shock so that, during the recovery period, the translation of housekeeping mRNAs can resume before the transcript processing machinery returns to normal. The posterior pole granule remains even more of a mystery but may be important for the metabolism of mRNPs that could be transported along to the plus end of cortical microtubules (Robinson et al. 1995). It has been shown that heat shock stress granules are localised to the periphery of the cell close to the subpellicular microtubules (Kramer et al. 2008). The heat shock response was present in the last common eukaryotic ancestor and trypanosomes subsequently lost the transcriptional component as they evolved to complete polycistronic transcription. One unique feature of trypanosome biology is the mechanisms that have evolved to compensate for the loss of the transcriptional response.
The unfolded protein response in mammalian cells
The unfolded protein response is induced when misfolded and unfolded proteins accumulate in the endoplasmic reticulum (ER). The UPR is activated by events that cause incomplete or unfolded proteins to accumulate—nutrient starvation, ER calcium depletion, hypoxia, reduction of disulphides by addition of dithiothreitol—and is also triggered by some viruses (Kohno 2009; Lee et al. 2009). The UPR is particularly apparent in secretory cells such as B-lymphocytes, pancreatic β cells and osteoblasts (Wu and Kaufman 2006). Although the UPR involves general down-regulation of mRNA translation to reduce the ER throughput of proteins, ER chaperones are selectively synthesised during the UPR.
Stress caused by unfolded proteins in the ER has to be transmitted to the cytoplasm. Three sensors have been characterised in mammalian cells: inositol-requiring enzyme 1 (IRE1), PKR-like ER kinase (PERK) and activating transcription factor 6 (ATF6). All are transmembrane proteins with a sensor domain located in the ER lumen and cytosolic domains transmitting the signal. In unstressed cells, all three of the sensor domains are associated with BiP, the predominant HSP70 homologue present in the ER. On accumulation of unfolded proteins in the ER, there is increased competition for BiP binding due to the unfolded proteins, and IRE1, PERK and ATF6 are activated, possibly by simple competition and/or an unknown signal (Bertolotti et al. 2000; Shen et al. 2002).
The loss of BiP binding results in IRE1 dimerisation and autophosphorylation that activates a nuclease domain that has two functions: (a) the degradation of mRNAs targeted to the ER (Hollien et al. 2009), thus reducing the input of newly synthesised polypeptides, and (b) cleavage of an intron in the XBP1 mRNA. The mRNA made by this cytoplasmic splicing encodes a transcription factor, pXBP1(S), required for the expression of UPR target genes (Yoshida et al. 2001; Uemura et al., 2009). The protein encoded by the unspliced mRNA, pXBP1(U), interacts with pXBP1(S) and sequesters it in the cytosol where the complex is rapidly degraded by the proteosome in response to a degradation motif in pXBP1(U) (Yoshida et al. 2006). Thus, pXBP1(U) is a feedback regulator of pXBP1(S) and effectively represses the transcription of UPR target genes during the recovery phase after ER stress.
Upon ER stress and release from BiP, PERK dimerises and autophosphorylation results in the activation of the kinase activity in the cytosolic domain. This kinase phosphorylates the translation initiation factor eIF2α, resulting in a reduced rate of initiation and thus a reduction in total protein synthesis by the mechanism described above (Shi et al. 1998b; Harding et al., 1999). Paradoxically, eIF2α phoshorylation enhances the translation of mRNAs with inhibitory short open reading frames (uORFs) upstream of the main initiation codon (Jackson et al. 2010); this category includes 45–50% of mammalian genes (Calvo et al. 2009). For example, ATF4 mRNA is preferentially translated upon ER stress and has two short uORFs; uORF1 facilitates ribosome scanning and re-initiation at downstream coding regions, whereas uORF2 normally inhibits the translation of ATF4 ORF. In unstressed cells, eIF2α-GTP is available and ribosomes reinitiate translation at uOFR2 rather than the ATF4 ORF. In stressed cells, less eIF2-GTP is available and ribosomes scan through uORF2 and initiate translation at the ATF4 initiation codon (Vattem and Wek 2004; Lu et al. 2004). ATF4 is a transcription factor that increases the transcription of genes encoding amino acid transporters, redox enzymes for promoting protein folding in the ER lumen and other transcription factors including XBP1. The up-regulation of XBP1 amplifies the UPR.
The normal interaction between BiP and ATF6 results in the retention of the complex in the ER. On dissociation, ATF6 is trafficked to the Golgi apparatus in COPII vesicles (Shen et al. 2002). The transmembrane domain of ATF6 is cleaved by two Golgi proteases there and the cytosolic ATF6 cleavage product translocates to the nucleus where it acts as a transcription factor for ER chaperone and XBP1 genes (Yoshida et al. 2000; Ye et al. 2000). pXBP1(U) is also a negative regulator of ATF6 and targets it for destruction by the proteasome (Yoshida et al. 2009).
All three sensors of ER stress respond in part by stimulating a transcriptional response to increase the expression of genes involved in protein folding in the ER.
The unfolded protein response in kinetoplastids
Treatment of bloodstream-form cells with 1 mM dithiothreitol to reduce disulphides or tunicamycin to inhibit N-linked glycosylation had very little effect on mRNA levels assessed using a microarray representing the membrane trafficking system, including BiP and protein disulphide isomerases (Koumandou et al. 2008). These data provide strong evidence for the absence of a transcriptional response that adjusts mRNA levels. In addition, no increase in BiP was detected by western blotting (Koumandou et al. 2008; Izquierdo et al. 2009). There are no immediately apparent homologues of IRE1 or XBP1 in the genome. Together these findings led to the model in which bloodstream form trypanosomes have an ER protein folding and quality control system that is not up-regulated in response to misfolded proteins. However, the absence of a transcriptional response in bloodstream-form trypanosomes does not indicate lack of an UPR, albeit atypical, and it is possible that the trypanosome UPR operates at the translational level. Three potential eIF2α kinases are present in the genome, all readily identified through sequence identity in the kinase domain (Moraes et al. 2007). One is probably the orthologue of GCN2 and may be involved in modulating protein synthesis in response to amino acid levels. However, the effector domains of the other two have no clear identity with the effector domains characterised in yeast and mammals. One of these two has been characterised and is localised to the endosomal compartment and the membrane of the flagellar pocket, the unique site of endocytosis in trypanosomatids. Thus, this kinase is unlikely to mediate an ER stress response but may be involved in sensing protein or nutrient transport (Moraes et al. 2007). One of the other eIF2α kinases could be activated upon ER stress, leading to phosphorylation of eIF2α and subsequent reduction of eIF2-GTP and translation; however, it has no transmembrane domain and activation would have to occur via an intermediate to transfer the signal across the ER membrane. Under these conditions, an increase in the translation of certain mRNAs could occur by the use of cap-independent initiation or short ORFs before the main ORF. Alternatively, regulatory mRNA-binding proteins could be modified upon ER stress and directly increase the translatability of certain mRNAs. The efficiency of mRNA translation could also be modified by changing the poly(A) status of some mRNAs as there is a correlation between poly(A) length and translatability (Beilharz and Preiss 2007; Lackner et al. 2007). This could be achieved by changing the recruitment of deadenylases via mRNA-binding proteins to a certain subset of mRNAs or by elongation of the poly(A) tails by cytosolic poly(A) polymerases. However, there has been little investigation of these potential mechanisms in trypanosomes.
Do bloodstream form trypanosomes need an unfolded protein response? It has been argued that one reason for this lack of regulation may be the protein flux through the ER. The bloodstream form trypanosome is unusual: 15% of total cellular protein is the variant surface glycoprotein (VSG) which represents ∼90% of the total flux through the ER. It has been estimated that only 30% of nascent VSG 221 attains the native state; the remainder is recognised to be the ER quality control system and is degraded by the proteosome (Field et al. 2007). The trypanosome has monoallelic expression of VSGs and switches the identity of the expressed VSG occasionally, usually as a result of a gene conversion event (reviewed in Schwede and Carrington 2010). It is likely that different VSGs fold at different rates, but the absence of a UPR would indicate that the trypanosome cannot compensate for slow-folding VSGs by increasing chaperone concentrations. In turn, this indicates that there may be a second system that coordinates the rate of cell growth with the rate of mature, folded VSG production. In procyclic forms, there is evidence for a change in the transcriptome in response to 4 mM dithiothreitol treatment; the change was similar to UPR in other eukaryotes over 1 h (Goldshmidt et al. 2010). However, both cases of 4 mM dithiothreitol used with procyclic forms and 1 mM dithiothreitol used with bloodstream forms in the experiments described above resulted in cell death and it remains to be determined whether the changes observed in procyclic forms were solely due to the accumulation of unfolded proteins in the ER.
In summary, in yeasts and metazoa, the stress responses described above are in part triggered by altered sequestration of heat shock proteins. The immediate responses act at all post-transcriptional levels and have evolved to slow or stop growth whilst repair occurs. In contrast, the slower transcriptional response has evolved to optimise the repair pathways. In trypanosomes, it is likely that the immediate responses are similar; heat shock does cause an inhibition of splicing and translation and a subsequent arrest in growth similar to other eukaryotes. However, it will be interesting to determine the mechanism that has replaced the longer-term transcriptional response in other eukaryotes.
Developmental regulation of gene expression
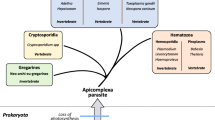
Trypanosomes proliferate in both vertebrate and invertebrate hosts. The cycle of transmission involves a series of developmentally distinct forms, at least ten in the case of T. brucei including four different forms that divide in four different host environments. Differences between the developmental forms include proliferation or arrest, morphology, cell surface architecture and metabolism (reviewed in Fenn and Matthews 2007; Sharma et al. 2009). Each form has a unique pattern of gene expression that is presumably determined by signals from the host, from other trypanosomes and by developmental history. As with the responses to stress described above, the vast majority of these changes occur in the absence of selective transcriptional control. However, there is an exception that probably represents a reappearance of transcriptional control in African trypanosomes; a small number of genes including those encoding two superabundant cell surface proteins are transcribed by RNAP I and are developmentally regulated by a suppression of transcription (reviewed in Rudenko 2010).
The molecular mechanisms that result in altered gene expression and differentiation in response to external signals may have a common evolutionary origin with one or more of the stress responses. In each case, there must be a sensor, a transduction pathway and then molecules that cause the response. In the stress responses discussed above, an example of a sensor is the interaction of BiP with ATF6; the interaction keeps the cell in an UPR-unstressed state whereas loss of interaction is characteristic of the stressed state. In the stress responses, the signal is both received and transduced by the proteins such as ATF6 to bring about changes in gene expression, transcriptional and post-transcriptional. Although the response to stress has been more or less well characterised in trypanosomes, the pathways that sense the stress and transduce the signal have not. Further, the differentiation processes in the trypanosome life cycle are more complex than a stress response: first, the transition is irreversible unlike the stress response. Second, the transition involves a more complex change in gene expression than the suppression of most genes that is apparent during stress. Third, most transitions from one developmental stage to the next involve a morphological change and, in most cases, this can only occur in coordination with a specialised cell cycle (Fig. 1). To expand this last point, the sleek elongated form of a trypanosome is produced by a spiralling array of microtubules present just inside the plasma membrane (Angelopoulos 1970); reviewed in Gull 1999). The microtubule array appears to be permanent and it accommodates cell growth through both lengthening of existing microtubules and the intercalation of new microtubules (Sherwin and Gull 1989). There is a single basal body that subtends the flagellum and is also physically connected to the kinetoplast (the single concatenated mass of mitochondrial DNA) through the mitochondrial membranes (Ogbadoyi et al. 2003). The point of emergence of the flagellum is the only discontinuity in the microtubule corset and the positioning of the hole through which the new daughter flagellum emerges is part of the coordinated construction of a new cytoskeleton that occurs throughout the growth phase of the cell division cycle (Robinson et al. 1995). Changes in cell length and the absolute and relative positions of the kinetoplast, which acts as an accurate marker for the basal body, and nucleus along the anterior–posterior axis accompany developmental transitions, for example, the nucleus can be positioned either anterior or posterior of the kinetoplast (reviewed in Sharma et al. 2009). During developmental transitions, some alterations in morphology occur as a result of lengthening of the microtubule corset. Other transitions are more complex, especially those that involve decreasing cell length and/or repositioning of the basal body along the anterior–posterior axis (Matthews et al. 1995). Such changes arise during a specialised asymmetric cell division that produces a daughter cell with a new morphology and the new basal body (Van Den Abbeele et al. 1999; Tyler et al. 2001b; Sharma et al. 2008) (Fig. 1). The major implication for the differentiation mechanism(s) is that the process has to be initiated before a discrete point in the cell cycle so that the new pattern of microtubule corset and thus cellular form is produced.
Cartoon showing the cellular differentiation processes occurring between the blood of a mammalian host and the tsetse fly midgut. The proliferative cell cycles are shown using dashed lines and the irreversible differentiation processes by solid lines. The known external signals for differentiation are shown in boxes. The predominant cell surface protein of each developmental stage is represented as follows: grey, VSG; green, GPEET procyclin; yellow, EP procyclin. The features of each cell are labelled in a representative cell in the top right section
It will be more informative to limit further discussion to two linked differentiation processes in trypanosomes that are characterised best: (a) the transition from slender, proliferative to stumpy, cell-cycle-arrested, bloodstream forms and (b) the differentiation of the stumpy bloodstream form to a proliferating procyclic form, similar to that present in the tsetse fly midgut (Fig. 1). Both transitions involve changes in cell morphology and gene expression (Roditi et al. 1989b; Dean et al. 2009; Tyler et al. 2001b; Matthews et al. 1995; Kabani et al. 2009). The discussion below is based partly on in vitro recapitulations but it is likely that it is a good reflection of in vivo events. In both cases, the external signal for differentiation is available and has provided the necessary tool to investigate events.
Slender to stumpy
Slender, bloodstream form trypanosomes can divide every 6 h and will overwhelm a host unless growth is curtailed. The trypanosome population is modulated both by the host response and by an intrinsic mechanism that limits density through differentiation to a non-proliferative, stumpy form that is unable to proliferate further unless ingested by the tsetse fly vector. The transition is not simply a cell division cycle arrest; stumpy cells have a different morphology, with a shorter flagellum, resulting from a specialised cell division cycle that produces two daughters, both of which become stumpy forms (Tyler et al. 2001b). Initially, the two daughters are not the same; the daughter with the new flagellum has stumpy morphology but the daughter with the old flagellum probably has to initiate flagellum shortening after division. It is unknown whether the daughter with the old flagellum always differentiates to a stumpy form or whether a fraction maintain slender morphology. Stumpy forms also express a set of genes not expressed in slender forms, notably those involved in metabolic pathways associated with the mitochondrion (Vickerman 1965; Tyler et al. 1997; Kabani et al. 2009)
The differentiation occurs in response to cell density, signalled by the production of stumpy induction factor (SIF) by trypanosomes (Vassella et al. 1997; Reuner et al. 1997; Tyler et al. 2001a). SIF was identified in conditioned medium and is a secreted molecule with a molecular weight of <500 but has not been defined further and the sensor is unknown. The signal transduction leading to differentiation to stumpy forms has been investigated; initial evidence suggested that a cAMP pathway was involved as membrane-permeable cAMP analogues induced differentiation as did etazolate, a phosphodiesterase inhibitor (Vassella et al. 1997). Subsequently, it was shown that membrane-permeable adenosine and AMP were more potent at inducing differentiation and it is likely that the active component is an intracellular metabolite of adenosine nucleotide as opposed to cAMP itself (Laxman et al. 2006). The induction of differentiation by etazolate is mysterious as it is not a potent inhibitor of the bulk phosphodiesterase activity in trypanosomes.
Some trypanosome cell lines do not differentiate from slender to stumpy forms, probably as a result of years of selection for rapid growth in laboratories. These cell lines produce SIF but do not respond, suggesting an absence of functional sensing or signalling and/or constitutive activation of a second pathway that overrides signalling from the SIF pathway. A pathway that maintains slender morphology and prevents arrest is evidenced by the deletion of genes encoding either MAP kinase 5 (MAPK5) or ZFK, a protein kinase with a novel domain architecture including a zinc finger. Deletion of either results in differentiation of slender to stumpy forms at a lower density than wild type, indicating that signalling through protein kinases is required to maintain the slender form (Domenicali Pfister et al. 2006; Vassella et al. 2001b). Thus, differentiation to stumpy forms could be triggered by a decrease in kinase activity and/or an increase in phosphatase activity.
Stumpy to procyclic
The transition from arrested stumpy cells to proliferating procyclic cells is the best-characterised trypanosome differentiation. The process can be recapitulated in culture and occurs as a series of ordered events. Changes in some mRNAs occur within the first hour; procyclins, the most abundant cell surface protein of procyclic forms, are expressed within 2 h and VSG, a major bloodstream form surface protein, is lost by 4–5 h, indicating an earlier loss of expression. The return to a proliferative cell division cycle and remodelling of the cell morphology has started by 6 h (Kabani et al. 2009).
The signals that induce differentiation have been characterised; both a decrease in temperature and the presence of the tricarboxylic acid cycle intermediate cis-aconitate are required (although a combination of cis-aconitate and citrate (CCA) is often used experimentally despite cis-aconitate itself is sufficient) (Ziegelbauer et al. 1990). The two signals interact; mammalian bloodstream-form trypanosomes are cultured at 37°C; insect procyclic forms at 27°C. If a bloodstream-form culture is transferred to a procyclic growth medium at 27°C, then the addition of a millimolar concentration of CCA is required (Ziegelbauer et al. 1990). However, if the temperature is reduced to 20°C, then only micromolar CCA is sufficient for full differentiation (Engstler and Boshart 2004). The drop in temperature to 20°C by itself is sufficient to induce the expression of the EP-1 procyclin, one of the superabundant cell surface proteins of procyclic forms, within 2 h (Engstler and Boshart 2004). However, the induction of EP-1 is reversible and the addition of CCA is required to induce the cellular differentiation processes, the change in cell morphology and the return to proliferation (Engstler and Boshart 2004). The most obvious change in morphology is the movement of the kinetoplast away from the posterior pole of the cell and closer to the nucleus and occurs during the first cell division cycle (Matthews et al. 1995).
Slender, proliferating bloodstream forms can differentiate efficiently to procyclic forms in vitro using the same signals (Czichos et al. 1986); however, the differentiation is asynchronous. The easiest explanation is that the cells are receptive to the differentiation signal at one point in the cell division cycle during G1, and stumpy forms are arrested at or just before this point whereas proliferative forms have to cycle to this point, resulting in asynchrony.
A second difference between slender and stumpy bloodstream form is the trafficking of cell surface proteins, first found for EP-1 procyclin. In slender forms it is not exported to the cell surface from the endosome, whereas in stumpy forms it is (Engstler and Boshart 2004). This difference in cell surface transport is important for the perception of the CCA signal that is sensed through regulated expression and cell surface trafficking of a tricarboxylic acid transporter encoded by the PAD2 gene (Dean et al. 2009). Decreasing the temperature from 37°C to 20°C is sufficient to both increase PAD2 protein levels and, in stumpy forms, change the subcellular localisation from the flagellar pocket to the entire plasma membrane. Depletion of PAD2 by RNAi diminishes both citrate uptake and differentiation to procyclics.
Entry of CCA into the cell breaks an autoregulatory cycle composed of two phosphatases, PTP and PIP39. The activity of PTP is necessary for maintaining the bloodstream-form state and inhibition or RNAi knockdown causes spontaneous differentiation of both slender and stumpy bloodstream to procyclic forms (Szoor et al. 2006). PIP39 interacts with and stimulates PTP activity. It has been proposed that the stimulatory activity is itself regulated by a balance between phosphorylation of PIP39 by an unknown kinase and dephosphorylation by PTP1 (Szoor et al. 2010). Entry of CCA into the cell breaks this cycle by decreasing PTP1 activity and thus causing an increase in phosphorylated PIP39. The phosphorylated PIP39 relocalises to the glycosome from where it drives the differentiation to procyclic forms. The finding that RNAi-mediated knockdown of PIP39 diminishes differentiation is support for this model.
The signalling process described above is by far the best-characterised response to a change in external environment in trypanosomes. The prospect of the identification of kinases and phosphatase substrates will eventually link the signalling pathway to the changes in gene expression.
Regulation of gene expression during developmental transitions
The importance of the identification of MAPK5, ZFK and the two phosphatases is that they provide solid evidence for control of differentiation of bloodstream to procyclic form by reversible phosphorylation. The downstream steps must be a change in gene expression and much of this change occurs via modulation of post-transcriptional processes. Details of how individual differentially expressed mRNAs are regulated are scarce, but it is becoming clear that instability of an individual mRNA in one life cycle stage but not in another is emerging as a theme for many developmentally regulated mRNAs and that the turnover of unstable mRNAs is dependent on the RNA helicase DHH1 (Kramer et al. 2010), the 5′–3′ exoribonuclease XRNA (Li et al. 2006) and the two deadenylases CAF1 and PAN2 (Schwede et al. 2009). What remains to be uncovered is the molecular mechanism that selects an individual mRNA to enter the instability pathway. The answer is going to be complex; DHH1 is a repressor of translation and the instability of differentially regulated mRNAs is intimately linked to whether they gain access to translation, a more complex process than translation-independent turnover.
Procyclins are the superabundant cell surface proteins of the procyclic form trypanosome and some details are known about the regulation of procyclin mRNAs. Procyclins, along with VSGs, are not typical protein-coding genes as they are transcribed by RNAP I (Johnson et al. 1987; Gunzl et al. 2003), with the 5′ cap being added by trans-splicing from a small RNAP II-transcribed exon (Sutton and Boothroyd 1986). It appears that only the VSGs and procyclins along with their associated co-transcribed genes are transcribed by RNAP I; the vast majority of genes are transcribed by RNAP II. The switch to transcription by RNAP I probably represents an adaptation to achieve high levels of expression; VSG is >10% of the total cell protein. The use of RNAP I is associated with the appearance of regulated transcription; procyclin transcription is reduced in bloodstream forms and VSGs are not transcribed in procyclic forms. Among the earliest detected changes during differentiation from bloodstream to procyclic forms are the suppression of VSG transcription and the loss of transcriptional suppression of the procyclin genes (Overath et al. 1983; Ehlers et al. 1987; Roditi et al. 1989a). These events are associated with structural changes in the nucleus as the VSG is transcribed from an extranucleolar focus of RNAP I, the expression site body, that disappears on differentiation (Navarro and Gull 2001; Landeira and Navarro 2007).
There are several procyclin isoforms that can be divided into two groups, GPEET and EP (Ruepp et al. 1997). Initially, both EP and GPEET procyclins are expressed within 2 h of a temperature decrease to 20°C, presumably corresponding to leaving the mammalian host (Ruepp et al. 1997; Engstler and Boshart 2004). After this initial burst of expression, GPEET procyclins predominate on the first ‘early’ procyclics that appear in the midgut within the peritrophic membrane. Subsequently, procyclic forms pass through the peritrophic membrane; around this time, GPEET expression is repressed and EP procyclin increases (Vassella et al. 2001a; Acosta-Serrano et al. 2001). The switch from GPEET to EP procyclins is reflected in the levels of the respective mRNAs; however, ‘late’ procyclics contain mRNAs for three EP procyclin isotypes, EP-1, EP-2 and EP-3, but only express two, EP-1 and EP-3. The mechanism and biological function of the translational repression of EP-2 are not understood.
The switch from GPEET to EP procyclin expression is linked to central metabolism, possibly reflecting carbon source availability (Vassella et al. 2000; Morris et al. 2002; Vassella et al. 2004). In culture, procyclic forms can use either substrate level phosphorylation or oxidative phosphorylation to generate ATP, the former predominating when glucose is available and the latter when glucose is depleted and the carbon source is amino acids, mainly proline (which is abundant in the tsetse midgut). During oxidative phosphorylation, NADH is not produced from the citric acid cycle but the acetate succinyl–coA transferase/succinyl–coA synthase cycle (van Weelden et al. 2005).
In ‘early’ procyclics in culture, GPEET expression can be maintained by the addition of glycerol to the medium (Vassella et al. 2000); thus, glycerol presents the switch to EP expression. In cultures of ‘late’ procyclics, predominantly expressing EP, knockdown of hexokinase or hexose transporters (Morris et al. 2002) or simply depletion of glucose from the medium resulted in increased GPEET and decreased EP expression; thus, a reduction in available glucose increases GPEET expression. The same effect occurs on suppression of mitochondrial catabolism by knockdown of either pyruvate dehydrogenase or succinyl CoA synthetase (Vassella et al. 2004). In contrast, inhibition of oxidative phosphorylation either through hypoxia or inhibition of the alternative oxidase suppresses GPEET mRNA. These manipulations of central metabolism presumably mimic a change in amount or type of carbon source between carbohydrates and amino acids, but it is difficult to make sense of the observations above by simply linking one carbon source or predominant catabolic pathway to the expression of either of procyclin types.
How is the carbon source state transmitted to mRNA levels? The finding that knockdown of subunits of AMP-dependent protein kinase resulted in the expression of both GPEET and EP procyclins in cells previously expressing just EP procyclin provides evidence for the identity of one component of the signalling pathway (Clemmens et al. 2009).
Despite the increase in transcription of procyclins on differentiation from bloodstream forms, much of the regulation of the procyclin mRNAs is post-transcriptional (Hug et al. 1994; Hehl et al. 1994; Hotz et al. 1997; Schürch et al. 1997). The 3′ UTRs contain elements that confer mRNA instability and translational suppression in bloodstream forms. Part of the EP-1 3′ UTR is necessary and sufficient for the induction of expression in bloodstream forms after dropping the temperature to 20°C as described above (Engstler and Boshart 2004). The GPEET 3′ UTR contains an element necessary for expression in response to glycerol (Vassella et al. 2000). Enormous efforts have gone into identifying trans-factors that interact with these elements; recent success has identified ZFP3, a zinc finger RNA-binding protein that selectively binds EP-1 and GPEET mRNAs that influences the balance between GPEET and EP-1 expression (Walrad et al. 2009).
The connection between factors involved in signalling differentiation and changing gene expression remains mysterious and the molecular characterisation of a complete pathway connecting an environmental change to alterations in mRNA stability or translation is a challenging goal. The evolutionary divergence of trypanosomes provides some information on the evolution of the response of gene expression to external signals. Trypanosomes represent a group that split at an early stage of the diversification of eukaryotes and it is likely that the heat shock response and ER unfolded protein response have evolved from pathways present in the last common eukaryotic ancestor and thus use common conserved genes such as heat shock proteins and MAP kinases. However, the differentiation processes probably evolved after divergence and the identities of components involved in signal sensing and possibly transduction are likely to vary. For example, yeast and metazoa frequently use receptors coupled to trimeric G-proteins to sense and transduce extracellular signals. Trimeric G-proteins are absent from the trypanosome genome and there are no apparent tyrosine kinase receptors. The identities of the sensor and transduction components in trypanosomes have and will emerge from both biochemical and post-genomic approaches. In particular, the recently developed forward genetics will provide answers.
Abbreviations
- RNAP:
-
RNA polymerase
- VSG:
-
Variant surface glycoprotein
- SL RNA:
-
Spliced leader RNA
- HSP:
-
Heat shock protein
References
Acosta-Serrano A, Vassella E, Liniger M, Kunz Renggli C, Brun R, Roditi I, Englund PT (2001) The surface coat of procyclic Trypanosoma brucei: programmed expression and proteolytic cleavage of procyclin in the tsetse fly. Proc Natl Acad Sci U S A 98:1513–1518
Anderson P, Kedersha N (2009) Stress granules. Curr Biol 19:R397–R398
Angelopoulos E (1970) Pellicular microtubules in the family Trypanosomatidae. J Protozool 17:39–51
Bangs JD, Crain PF, Hashizume T, McCloskey JA, Boothroyd JD (1992) Mass spectroscopy of mRNA cap4 from trypanosomatids reveals two novel nucleosides. J Biol Chem 267:9805–9815
Beilharz TH, Preiss T (2007) Widespread use of poly(A) tail length control to accentuate expression of the yeast transcriptome. RNA 13:982–997
Berriman M, Ghedin E, Hertz-Fowler C, Blandin G, Renauld H, Bartholomeu DC, Lennard NJ, Caler E, Hamlin NE, Haas B, Bohme U, Hannick L, Aslett MA, Shallom J, Marcello L, Hou L, Wickstead B, Alsmark UC, Arrowsmith C, Atkin RJ, Barron AJ, Bringaud F, Brooks K, Carrington M, Cherevach I, Chillingworth TJ, Churcher C, Clark LN, Corton CH, Cronin A, Davies RM, Doggett J, Djikeng A, Feldblyum T, Field MC, Fraser A, Goodhead I, Hance Z, Harper D, Harris BR, Hauser H, Hostetler J, Ivens A, Jagels K, Johnson D, Johnson J, Jones K, Kerhornou AX, Koo H, Larke N, Landfear S, Larkin C, Leech V, Line A, Lord A, Macleod A, Mooney PJ, Moule S, Martin DM, Morgan GW, Mungall K, Norbertczak H, Ormond D, Pai G, Peacock CS, Peterson J, Quail MA, Rabbinowitsch E, Rajandream MA, Reitter C, Salzberg SL, Sanders M, Schobel S, Sharp S, Simmonds M, Simpson AJ, Tallon L, Turner CM, Tait A, Tivey AR, Van Aken S, Walker D, Wanless D, Wang S, White B, White O, Whitehead S, Woodward J, Wortman J, Adams MD, Embley TM, Gull K, Ullu E, Barry JD, Fairlamb AH, Opperdoes F, Barrell BG, Donelson JE, Hall N, Fraser CM, Melville SE, El-Sayed NM (2005) The genome of the African trypanosome Trypanosoma brucei. Science 309:416–422
Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D (2000) Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol 2:326–332
Calvo SE, Pagliarini DJ, Mootha VK (2009) Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc Natl Acad Sci U S A 106:7507–7512
Clayton CE, Shapira M (2007) Post-transcriptional regulation of gene expression in trypanosomes and leishmanias. Mol Biochem Parasitol 156:93–101
Clemmens CS, Morris MT, Lyda TA, Acosta-Serrano A, Morris JC (2009) Trypanosoma brucei AMP-activated kinase subunit homologs influence surface molecule expression. Exp Parasitol 123:250–257
Clos J, Westwood JT, Becker PB, Wilson S, Lambert K, Wu C (1990) Molecular cloning and expression of a hexameric Drosophila heat shock factor subject to negative regulation. Cell 63:1085–1097
Czichos J, Nonnengaesser C, Overath P (1986) Trypanosoma brucei: cis-aconitate and temperature reduction as triggers of synchronous transformation of bloodstream to procyclic trypomastigotes in vitro. Exp Parasitol 62:283–291
Dacks JB, Walker G, Field MC (2008) Implications of the new eukaryotic systematics for parasitologists. Parasitol Int 57:97–104
Daulny A, Geng F, Muratani M, Geisinger JM, Salghetti SE, Tansey WP (2008) Modulation of RNA polymerase II subunit composition by ubiquitylation. Proc Natl Acad Sci U S A 105:19649–19654
David M, Gabdank I, Ben-David M, Zilka A, Orr I, Barash D, Shapira M (2009) Preferential translation of Hsp83 in Leishmania requires a thermosensitive polypyrimidine-rich element in the 3′ UTR and involves scanning of the 5′ UTR. RNA 16:364–374
Dean S, Marchetti R, Kirk K, Matthews KR (2009) A surface transporter family conveys the trypanosome differentiation signal. Nature 459:213–217
Domenicali Pfister D, Burkard G, Morand S, Renggli CK, Roditi I, Vassella E (2006) A mitogen-activated protein kinase controls differentiation of bloodstream forms of Trypanosoma brucei. Eukaryot Cell 5:1126–1135
Edwards AM, Kane CM, Young RA, Kornberg RD (1991) Two dissociable subunits of yeast RNA polymerase II stimulate the initiation of transcription at a promoter in vitro. J Biol Chem 266:71–75
Ehlers B, Czichos J, Overath P (1987) Repression and reactivation of the variant surface glycoprotein gene in Trypanosoma brucei. FEBS Lett 225:53–58
Emara MM, Ivanov P, Hickman T, Dawra N, Tisdale S, Kedersha N, Hu GF, Anderson P (2010) Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J Biol Chem 285:10959–10968
Engstler M, Boshart M (2004) Cold shock and regulation of surface protein trafficking convey sensitization to inducers of stage differentiation in Trypanosoma brucei. Genes Dev 18:2798–2811
Ersfeld K, Asbeck K, Gull K (1998) Direct visualisation of individual gene organisation in Trypanosoma brucei by high-resolution in situ hybridisation. Chromosoma 107:237–240
Farny NG, Kedersha NL, Silver PA (2009) Metazoan stress granule assembly is mediated by P-eIF2alpha-dependent and -independent mechanisms. RNA 15:1814–1821
Fenn K, Matthews KR (2007) The cell biology of Trypanosoma brucei differentiation. Curr Opin Microbiol 10:539–546
Field MC, Sergeenko T, Wang YN, Bohm S, Carrington M (2007) Chaperone requirements for biosynthesis of the trypanosome variant surface glycoprotein. PLoS One 5:e8468
Folgueira C, Requena JM (2007) Pitfalls of the CAT reporter gene for analyzing translational regulation in Leishmania. Parasitol Res 101:1449–1452
Folgueira C, Quijada L, Soto M, Abanades DR, Alonso C, Requena JM (2005) The translational efficiencies of the two Leishmania infantum HSP70 mRNAs, differing in their 3′-untranslated regions, are affected by shifts in the temperature of growth through different mechanisms. J Biol Chem 280:35172–35183
Gallouzi IE, Brennan CM, Stenberg MG, Swanson MS, Eversole A, Maizels N, Steitz JA (2000) HuR binding to cytoplasmic mRNA is perturbed by heat shock. Proc Natl Acad Sci U S A 97:3073–3078
Garcia-Silva MR, Frugier M, Tosar JP, Correa-Dominguez A, Romalto-Alves L, Parodi-Talice A, Rovira C, Robello C, Goldenberg S, Cayota A (2010) A population of tRNA-derived small RNAs is actively produced in Trypanosoma cruzi and recruited to specific cytoplasmic granules. Mol Biochem Parasitol 171:64–73
Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO (2000) Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11:4241–4257
Gilinger G, Bellofatto V (2001) Trypanosome spliced leader RNA genes contain the first identified RNA polymerase II gene promoter in these organisms. Nucl Acids Res 29:1556–1564
Goldshmidt H, Matas D, Kabi A, Carmi S, Hope R, Michaeli S (2010) Persistent ER stress induces the spliced leader RNA silencing pathway (SLS), leading to programmed cell death in Trypanosoma brucei. PLoS Pathog 6:e1000731
Goler-Baron V, Selitrennik M, Barkai O, Haimovich G, Lotan R, Choder M (2008) Transcription in the nucleus and mRNA decay in the cytoplasm are coupled processes. Genes Dev 22:2022–2027
Grousl T, Ivanov P, Frydlova I, Vasicova P, Janda F, Vojtova J, Malinska K, Malcova I, Novakova L, Janoskova D, Valasek L, Hasek J (2009) Robust heat shock induces eIF2alpha-phosphorylation-independent assembly of stress granules containing eIF3 and 40S ribosomal subunits in budding yeast, Saccharomyces cerevisiae. J Cell Sci 122:2078–2088
Guettouche T, Boellmann F, Lane WS, Voellmy R (2005) Analysis of phosphorylation of human heat shock factor 1 in cells experiencing a stress. BMC Biochem 6:4
Gull K (1999) The cytoskeleton of trypanosomatid parasites. Annu Rev Microbiol 53:629–655
Gunzl A, Bruderer T, Laufer G, Schimanski B, Tu LC, Chung HM, Lee PT, Lee MG (2003) RNA polymerase I transcribes procyclin genes and variant surface glycoprotein gene expression sites in Trypanosoma brucei. Eukaryot Cell 2:542–551
Haanstra J, Stewart M, Luu V-D, van Tuijl A, Westerhoff H, Clayton CE, Bakker B (2008) Control and regulation of gene expression: quantitative analysis of the expression of phosphoglycerate kinase in bloodstream form Trypanosoma brucei. J Biol Chem 283:2495–2507
Harding HP, Zhang Y, Ron D (1999) Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397:271–274
Harel-Sharvit L, Eldad N, Haimovich G, Barkai O, Duek L, Choder M (2010) RNA polymerase II subunits link transcription and mRNA decay to translation. Cell 143:552–563
Hausler T, Clayton C (1996) Post-transcriptional control of hsp70 mRNA in Trypanosoma brucei. Mol Biochem Parasitol 76:57–71
Hehl A, Vassella E, Braun R, Roditi I (1994) A conserved stem-loop structure in the 3′ untranslated region of procyclin mRNAs regulates expression in Trypanosoma brucei. Proc Natl Acad Sci U S A 91:370–374
Hietakangas V, Anckar J, Blomster HA, Fujimoto M, Palvimo JJ, Nakai A, Sistonen L (2006) PDSM, a motif for phosphorylation-dependent SUMO modification. Proc Natl Acad Sci U S A 103:45–50
Hollien J, Lin JH, Li H, Stevens N, Walter P, Weissman JS (2009) Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J Cell Biol 186:323–331
Holmberg CI, Hietakangas V, Mikhailov A, Rantanen JO, Kallio M, Meinander A, Hellman J, Morrice N, MacKintosh C, Morimoto RI, Eriksson JE, Sistonen L (2001) Phosphorylation of serine 230 promotes inducible transcriptional activity of heat shock factor 1. EMBO J 20:3800–3810
Hong Y, Rogers R, Matunis MJ, Mayhew CN, Goodson ML, Park-Sarge OK, Sarge KD (2001) Regulation of heat shock transcription factor 1 by stress-induced SUMO-1 modification. J Biol Chem 276:40263–40267
Hotz HR, Hartmann C, Huober K, Hug M, Clayton C (1997) Mechanisms of developmental regulation in Trypanosoma brucei: a polypyrimidine tract in the 3′-untranslated region of a surface protein mRNA affects RNA abundance and translation. Nucleic Acids Res 15:3017–3025
Hug M, Hotz HR, Hartmann C, Clayton C (1994) Hierarchies of RNA-processing signals in a trypanosome surface antigen mRNA precursor. Mol Cell Biol 14:7428–7435
Izquierdo L, Atrih A, Rodrigues JA, Jones DC, Ferguson MA (2009) Trypanosoma brucei UDP-glucose:glycoprotein glucosyltransferase has unusual substrate specificity and protects the parasite from stress. Eukaryot Cell 8:230–240
Jackson RJ, Hellen CU, Pestova TV (2010) The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11:113–127
Jae N, Wang P, Gu T, Huhn M, Palfi Z, Urlaub H, Bindereif A (2010) Essential role of a trypanosome U4-specific Sm core protein in small nuclear ribonucleoprotein assembly and splicing. Eukaryot Cell 9:379–386
Johnson PJ, Kooter JM, Borst P (1987) Inactivation of transcription by UV irradiation of T. brucei provides evidence for a multicistronic transcription unit including a VSG gene. Cell 51:273–281
Kabani S, Fenn K, Ross A, Ivens A, Smith TK, Ghazal P, Matthews K (2009) Genome-wide expression profiling of in vivo-derived bloodstream parasite stages and dynamic analysis of mRNA alterations during synchronous differentiation in Trypanosoma brucei. BMC Genomics 10:427
Kedersha N, Anderson P (2002) Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem Soc Trans 30:963–969
Kedersha NL, Gupta M, Li W, Miller I, Anderson P (1999) RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2a to the assembly of mammalian stress granules. J Cell Biol 147:1431–1441
Kim SA, Yoon JH, Lee SH, Ahn SG (2005) Polo-like kinase 1 phosphorylates heat shock transcription factor 1 and mediates its nuclear translocation during heat stress. J Biol Chem 280:12653–12657
Kline MP, Morimoto RI (1997) Repression of the heat shock factor 1 transcriptional activation domain is modulated by constitutive phosphorylation. Mol Cell Biol 17:2107–2115
Knauf U, Newton EM, Kyriakis J, Kingston RE (1996) Repression of human heat shock factor 1 activity at control temperature by phosphorylation. Genes Dev 10:2782–2793
Kohno K (2009) Stress-sensing mechanisms in the unfolded protein response: similarities and differences between yeast and mammals. J Biochem 147:27–33
Kolev NG, Franklin JB, Carmi S, Shi H, Michaeli S, Tschudi C (2010) The transcriptome of the human pathogen Trypanosoma brucei at single-nucleotide resolution. PLOS Pathogens 6:e1001090
Koumandou VL, Natesan SK, Sergeenko T, Field MC (2008) The trypanosome transcriptome is remodelled during differentiation but displays limited responsiveness within life stages. BMC Genomics 9:298
Kramer S, Queiroz R, Ellis L, Webb H, Hoheisel JD, Clayton C, Carrington M (2008) Heat shock causes a decrease in polysomes and the appearance of stress granules in trypanosomes independently of eIF2(alpha) phosphorylation at Thr169. J Cell Sci 121:3002–3014
Kramer S, Queiroz R, Ellis L, Hoheisel JD, Clayton C, Carrington M (2010) The RNA helicase DHH1 is central to the correct expression of many developmentally regulated mRNAs in trypanosomes. J Cell Sci 123:699–711
Lackner DH, Beilharz TH, Marguerat S, Mata J, Watt S, Schubert F, Preiss T, Bahler J (2007) A network of multiple regulatory layers shapes gene expression in fission yeast. Mol Cell 26:145–155
Landeira D, Navarro M (2007) Nuclear repositioning of the VSG promoter during developmental silencing in Trypanosoma brucei. J Cell Biol 176:133–139
Laroia G, Cuesta R, Brewer G, Schneider RJ (1999) Control of mRNA decay by heat shock-ubiquitin-proteasome pathway. Science 284:499–502
Larreta R, Soto M, Quijada L, Folgueira C, Abanades DR, Alonso C, Requena JM (2004) The expression of HSP83 genes in Leishmania infantum is affected by temperature and by stage-differentiation and is regulated at the levels of mRNA stability and translation. BMC Mol Biol 5:3
Laxman S, Riechers A, Sadilek M, Schwede F, Beavo JA (2006) Hydrolysis products of cAMP analogs cause transformation of Trypanosoma brucei from slender to stumpy-like forms. Proc Natl Acad Sci U S A 103:19194–19199
LeBowitz JH, Smith HQ, Rusche L, Beverley SM (1993) Coupling of poly(A) site selection and trans-splicing in Leishmania. Genes Dev 7:996–1007
Lee MG (1995) Heat shock does not increase the transcriptional efficiency of the Hsp 70 genes of Trypanosoma brucei. Exp Parasitol 81:608–613
Lee MG (1998) The 3′ untranslated region of the hsp 70 genes maintains the level of steady state mRNA in Trypanosoma brucei upon heat shock. Nucleic Acids Res 26:4025–4033
Lee DY, Lee J, Sugden B (2009) The unfolded protein response and autophagy: herpesviruses rule! J Virol 83:1168–1172
Li CH, Irmer H, Gudjonsdottir-Planck D, Freese S, Salm H, Haile S, Estevez AM, Clayton C (2006) Roles of a Trypanosoma brucei 5′->3′ exoribonuclease homolog in mRNA degradation. RNA 12:2171–2186
Lotan R, Bar-On VG, Harel-Sharvit L, Duek L, Melamed D, Choder M (2005) The RNA polymerase II subunit Rpb4p mediates decay of a specific class of mRNAs. Genes Dev 19:3004–3016
Lotan R, Goler-Baron V, Duek L, Haimovich G, Choder M (2007) The Rpb7p subunit of yeast RNA polymerase II plays roles in the two major cytoplasmic mRNA decay mechanisms. J Cell Biol 178:1133–1143
Lu PD, Harding HP, Ron D (2004) Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J Cell Biol 167:27–33
Mair G, Shi H, Li H, Djikeng A, Aviles HO, Bishop JR, Falcone FH, Gavrilescu C, Montgomery JL, Santori MI, Stern LS, Wang Z, Ullu E, Tschudi C (2000) A new twist in trypanosome RNA metabolism: cis-splicing of pre-mRNA. RNA 6:163–169
Martin F, Requena JM, Martin J, Alonso C, Lopez MC (1993) Cytoplasmic–nuclear translocation of the Hsp70 protein during environmental stress in Trypanosoma cruzi. Biochem Biophys Res Commun 196:1155–1162
Martinez-Calvillo S, Yan S, Nguyen D, Fox M, Stuart K, Myler PJ (2003) Transcription of Leishmania major Friedlin chromosome 1 initiates in both directions within a single region. Mol Cell 11:1291–1299
Martinez-Calvillo S, Nguyen D, Stuart K, Myler PJ (2004) Transcription initiation and termination on Leishmania major chromosome 3. Eukaryot Cell 3:506–517
Matthews KR, Tschudi C, Ullu E (1994) A common pyrimidine-rich motif governs trans-splicing and polyadenylation of tubulin polycistronic pre-mRNA in trypanosomes. Genes Dev 8:491–501
Matthews KR, Sherwin T, Gull K (1995) Mitochondrial genome repositioning during the differentiation of the African trypanosome between life cycle forms is microtubule mediated. J Cell Sci 108:2231–2239
McEwen E, Kedersha N, Song B, Scheuner D, Gilks N, Han A, Chen JJ, Anderson P, Kaufman RJ (2005) Heme-regulated inhibitor kinase-mediated phosphorylation of eukaryotic translation initiation factor 2 inhibits translation, induces stress granule formation, and mediates survival upon arsenite exposure. J Biol Chem 280:16925–16933
Moraes MC, Jesus TC, Hashimoto NN, Dey M, Schwartz KJ, Alves VS, Avila CC, Bangs JD, Dever TE, Schenkman S, Castilho BA (2007) Novel membrane-bound eIF2alpha kinase in the flagellar pocket of Trypanosoma brucei. Eukaryot Cell 6:1979–1991
Morris JC, Wang Z, Drew ME, Englund PT (2002) Glycolysis modulates trypanosome glycoprotein expression as revealed by an RNAi library. EMBO J 21:4429–4438
Muhich ML, Boothroyd JC (1988) Polycistronic transcripts in trypanosomes and their accumulation during heat shock: evidence for a precursor role in mRNA synthesis. Mol Cell Biol 8:3837–3846
Muhich ML, Boothroyd JC (1989) Synthesis of trypanosome HSP70 mRNA is resistant to disruption of trans-splicing by heat shock. J Biol Chem 264:7107–7110
Muhlrad D, Parker R (1999) Recognition of yeast mRNAs as “nonsense containing” leads to both inhibition of mRNA translation and mRNA degradation: implications for the control of mRNA decapping. Mol Biol Cell 10:3971–3978
Navarro M, Gull K (2001) A pol I transcriptional body associated with VSG mono-allelic expression in Trypanosoma brucei. Nature 414:759–763
Nilsson D, Gunasekera K, Mani J, Osteras M, Farinelli L, Baerlocher L, Roditi I, Ochsenreiter T (2010) Spliced leader trapping reveals widespread alternative splicing patterns in the highly dynamic transcriptome of Trypanosoma brucei. PLoS Pathog 6:e1001037
Ogbadoyi EO, Robinson DR, Gull K (2003) A high-order trans-membrane structural linkage is responsible for mitochondrial genome positioning and segregation by flagellar basal bodies in trypanosomes. Mol Biol Cell 14:1769–1779
Olson CL, Nadeau KC, Sullivan MA, Winquist AG, Donelson JE, Walsh CT, Engman DM (1994) Molecular and biochemical comparison of the 70-kDa heat shock proteins of Trypanosoma cruzi. J Biol Chem 269:3868–3874
Orlicky SM, Tran PT, Sayre MH, Edwards AM (2001) Dissociable Rpb4–Rpb7 subassembly of RNA polymerase II binds to single-strand nucleic acid and mediates a post-recruitment step in transcription initiation. J Biol Chem 276:10097–10102
Overath P, Czichos J, Stock U, Nonnengaesser C (1983) Repression of glycoprotein synthesis and release of surface coat during transformation of Trypanosoma brucei. EMBO J 2:1721–1728
Pirkkala L, Nykanen P, Sistonen L (2001) Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB J 15(7):1118–1131
Quijada L, Soto M, Alonso C, Requena JM (1997) Analysis of post-transcriptional regulation operating on transcription products of the tandemly linked Leishmania infantum hsp70 genes. J Biol Chem 272:4493–4499
Quijada L, Soto M, Alonso C, Requena JM (2000) Identification of a putative regulatory element in the 3′-untranslated region that controls expression of HSP70 in Leishmania infantum. Mol Biochem Parasitol 110:79–91
Reuner B, Vassella E, Yutzy B, Boshart M (1997) Cell density triggers slender to stumpy differentiation of Trypanosoma brucei bloodstream forms in culture. Mol Biochem Parasitol 90:269–280
Robinson DR, Sherwin T, Ploubidou A, Byard EH, Gull K (1995) Microtubule polarity and dynamics in the control of organelle positioning, segregation, and cytokinesis in the trypanosome cell cycle. J Cell Biol 128:1163–1172
Roditi I, Schwartz H, Pearson TW, Beecroft RP, Liu MK, Richardson JP, Bühring H-J, Pleiss J, Bülow R, Williams RO, Overath P (1989a) Procyclin gene expression and loss of the variant surface glycoprotein during differentiation of Trypanosoma brucei. J Cell Biol 108:737–746
Roditi I, Schwarz H, Pearson TW, Beecroft RP, Liu MK, Richardson JP, Buhring HJ, Pleiss J, Bulow R, Williams RO et al (1989b) Procyclin gene expression and loss of the variant surface glycoprotein during differentiation of Trypanosoma brucei. J Cell Biol 108:737–746
Rubtsova MP, Sizova DV, Dmitriev SE, Ivanov DS, Prassolov VS, Shatsky IN (2003) Distinctive properties of the 5′-untranslated region of human hsp70 mRNA. J Biol Chem 278:22350–22356
Rudenko G (2010) Epigenetics and transcriptional control in African trypanosomes. Essays Biochem 48:201–219
Ruepp S, Furger A, Kurath U, Renggli CK, Hemphill A, Brun R, Roditi I (1997) Survival of Trypanosoma brucei in the tsetse fly is enhanced by the expression of specific forms of procyclin. J Cell Biol 137:1369–1379
Runner VM, Podolny V, Buratowski S (2008) The Rpb4 subunit of RNA polymerase II contributes to cotranscriptional recruitment of 3′ processing factors. Mol Cell Biol 28:1883–1891
Saavedra C, Tung KS, Amberg DC, Hopper AK, Cole CN (1996) Regulation of mRNA export in response to stress in Saccharomyces cerevisiae. Genes Dev 10:1608–1620
Sarge KD, Murphy SP, Morimoto RI (1993) Activation of heat shock gene transcription by heat shock factor 1 involves oligomerization, acquisition of DNA-binding activity, and nuclear localization and can occur in the absence of stress. Mol Cell Biol 13:1392–1407
Schürch N, Furger A, Kurath U, Roditi I (1997) Contributions of the procyclin 3′ untranslated region and coding region to the regulation of expression in bloodstream forms of Trypanosoma brucei. Mol Biochem Parasitol 89:109–121
Schwede A, Carrington M (2010) Bloodstream form trypanosome plasma membrane proteins: antigenic variation and invariant antigens. Parasitology 137:1–11
Schwede A, Ellis L, Luther J, Carrington M, Stoecklin G, Clayton C (2008) A role for Caf1 in mRNA deadenylation and decay in trypanosomes and human cells. Nucleic Acids Res 36:3374–3388
Schwede A, Manful T, Jha BA, Helbig C, Bercovich N, Stewart M, Clayton C (2009) The role of deadenylation in the degradation of unstable mRNAs in trypanosomes. Nucleic Acids Res 37:5511–5528
Selitrennik M, Duek L, Lotan R, Choder M (2006) Nucleocytoplasmic shuttling of the Rpb4p and Rpb7p subunits of Saccharomyces cerevisiae RNA polymerase II by two pathways. Eukaryot Cell 5:2092–2103
Sharma R, Peacock L, Gluenz E, Gull K, Gibson W, Carrington M (2008) Asymmetric cell division as a route to reduction in cell length and change in cell morphology in trypanosomes. Protist 159:137–151
Sharma R, Gluenz E, Peacock L, Gibson W, Gull K, Carrington M (2009) The heart of darkness: growth and form of Trypanosoma brucei in the tsetse fly. Trends Parasitol 25:517–524
Shen J, Chen X, Hendershot L, Prywes R (2002) ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev Cell 3:99–111
Sherwin T, Gull K (1989) Visualization of detyrosination along single microtubules reveals novel mechanisms of assembly during cytoskeletal duplication in trypanosomes. Cell 57:211–221
Shi Y, Mosser DD, Morimoto RI (1998a) Molecular chaperones as HSF1-specific transcriptional repressors. Genes Dev 12:654–666
Shi Y, Vattem KM, Sood R, An J, Liang J, Stramm L, Wek RC (1998b) Identification and characterization of pancreatic eukaryotic initiation factor 2 alpha-subunit kinase, PEK, involved in translational control. Mol Cell Biol 18:7499–7509
Shin C, Feng Y, Manley JL (2004) Dephosphorylated SRp38 acts as a splicing repressor in response to heat shock. Nature 427:553–558
Siegel TN, Tan KSW, Cross GAM (2005) Systematic study of sequence motifs for RNA trans splicing in Trypanosoma brucei. Mol Cell Biol 25:9586–9594
Siegel TN, Hekstra DR, Kemp LA, Figuiredo LM, Lowell JE, Fenyo D, Wang X, Dewell S, Coross GAM (2009) Four histone variants mark the boundaries of polycistronic transcription units in Trypanosoma brucei. Genes Dev 23:1063–1076
Siegel TN, Hekstra DR, Wang X, Dewell S, Cross GA (2010) Genome-wide analysis of mRNA abundance in two life-cycle stages of Trypanosoma brucei and identification of splicing and polyadenylation sites. Nucleic Acids Res 38:4946–4957
Sorger PK, Pelham HR (1988) Yeast heat shock factor is an essential DNA-binding protein that exhibits temperature-dependent phosphorylation. Cell 54:855–864
Stohr N, Lederer M, Reinke C, Meyer S, Hatzfeld M, Singer RH, Huttelmaier S (2006) ZBP1 regulates mRNA stability during cellular stress. J Cell Biol 175:527–534
Sutton RE, Boothroyd JC (1986) Evidence for trans splicing in trypanosome. Cell 47:527–535
Szoor B, Wilson J, McElhinney H, Tabernero L, Matthews KR (2006) Protein tyrosine phosphatase TbPTP1: a molecular switch controlling life cycle differentiation in trypanosomes. J Cell Biol 175:293–303
Szoor B, Ruberto I, Burchmore R, Matthews KR (2010) A novel phosphatase cascade regulates differentiation in Trypanosoma brucei via a glycosomal signaling pathway. Genes Dev 24:1306–1316
Theodorakis NG, Morimoto RI (1987) Post-transcriptional regulation of hsp70 expression in human cells: effects of heat shock, inhibition of protein synthesis, and adenovirus infection on translation and mRNA stability. Mol Cell Biol 7:4357–4368
Thompson DM, Parker R (2009) The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae. J Cell Biol 185:43–50
Tyler KM, Matthews KR, Gull K (1997) The bloodstream differentiation–division of Trypanosoma brucei studied using mitochondrial markers. Proc Biol Sci 264:1481–1490
Tyler KM, Higgs PG, Matthews KR, Gull K (2001a) Limitation of Trypanosoma brucei parasitaemia results from density-dependent parasite differentiation and parasite killing by the host immune response. Proc Biol Sci 268:2235–2243
Tyler KM, Matthews KR, Gull K (2001b) Anisomorphic cell division by African trypanosomes. Protist 152:367–378
Uemura A, Oku M, Mori K, Yoshida H (2009) Unconventional splicing of XBP1 mRNA occurs in the cytoplasm during the mammalian unfolded protein response. J Cell Sci 122:2877–2886
Ullu E, Matthews KR, Tschudi C (1993) Temporal order of RNA-processing reactions in trypanosomes: rapid trans splicing precedes polyadenylation of newly synthesised tubulin transcripts. Mol Cell Biol 13:720–725
Van Den Abbeele J, Claes Y, van Bockstaele D, Le Ray D, Coosemans M (1999) Trypanosoma brucei spp. development in the tsetse fly: characterization of the post-mesocyclic stages in the foregut and proboscis. Parasitology 118:469–478
van Weelden SW, van Hellemond JJ, Opperdoes FR, Tielens AG (2005) New functions for parts of the Krebs cycle in procyclic Trypanosoma brucei, a cycle not operating as a cycle. J Biol Chem 280:12451–12460
Vassella E, Reuner B, Yutzy B, Boshart M (1997) Differentiation of African trypanosomes is controlled by a density sensing mechanism which signals cell cycle arrest via the cAMP pathway. J Cell Sci 110:2661–2671
Vassella E, Den Abbeele JV, Butikofer P, Renggli CK, Furger A, Brun R, Roditi I (2000) A major surface glycoprotein of Trypanosoma brucei is expressed transiently during development and can be regulated post-transcriptionally by glycerol or hypoxia. Genes Dev 14:615–626
Vassella E, Acosta-Serrano A, Studer E, Lee SH, Englund PT, Roditi I (2001a) Multiple procyclin isoforms are expressed differentially during the development of insect forms of Trypanosoma brucei. J Mol Biol 312:597–607
Vassella E, Kramer R, Turner CM, Wankell M, Modes C, van den Bogaard M, Boshart M (2001b) Deletion of a novel protein kinase with PX and FYVE-related domains increases the rate of differentiation of Trypanosoma brucei. Mol Microbiol 41:33–46
Vassella E, Probst M, Schneider A, Studer E, Renggli CK, Roditi I (2004) Expression of a major surface protein of Trypanosoma brucei insect forms is controlled by the activity of mitochondrial enzymes. Mol Biol Cell 15:3986–3993
Vattem KM, Wek RC (2004) Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc Natl Acad Sci U S A 101(31):11269–11274
Vickerman K (1965) Polymorphism and mitochondrial activity in sleeping sickness trypanosomes. Nature 208:762–766
von Koskull-Doring P, Scharf KD, Nover L (2007) The diversity of plant heat stress transcription factors. Trends Plant Sci 12:452–457
Walrad P, Paterou A, Acosta-Serrano A, Matthews KR (2009) Differential trypanosome surface coat regulation by a CCCH protein that co-associates with procyclin mRNA cis-elements. PLoS Pathog 5:e1000317
Webb H, Burns R, Ellis L, Kimblin N, Carrington M (2005) Developmentally regulated instability of the GPI-PLC mRNA is dependent on a short lived protein factor. Nucleic Acids Res 33:1503–1512
Welch WJ, Suhan JP (1985) Morphological study of the mammalian stress response: characterization of changes in cytoplasmic organelles, cytoskeleton, and nucleoli, and appearance of intranuclear actin filaments in rat fibroblasts after heat-shock treatment. J Cell Biol 101:1198–1211
Wu J, Kaufman RJ (2006) From acute ER stress to physiological roles of the unfolded protein response. Cell Death Differ 13:374–384
Ye J, Rawson RB, Komuro R, Chen X, Dave UP, Prywes R, Brown MS, Goldstein JL (2000) ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell 6:1355–1364
Yoshida H, Okada T, Haze K, Yanagi H, Yura T, Negishi M, Mori K (2000) ATF6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cis-acting element responsible for the mammalian unfolded protein response. Mol Cell Biol 20:6755–6767
Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K (2001) XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107:881–891
Yoshida H, Oku M, Suzuki M, Mori K (2006) pXBP1(U) encoded in XBP1 pre-mRNA negatively regulates unfolded protein response activator pXBP1(S) in mammalian ER stress response. J Cell Biol 172:565–575
Yoshida H, Uemura A, Mori K (2009) pXBP1(U), a negative regulator of the unfolded protein response activator pXBP1(S), targets ATF6 but not ATF4 in proteasome-mediated degradation. Cell Struct Funct 34:1–10
Yost HJ, Lindquist S (1991) Heat shock proteins affect RNA processing during the heat shock response of Saccharomyces cerevisiae. Mol Cell Biol 11:1062–1068
Ziegelbauer K, Quinten M, Schwarz H, Pearson TW, Overath P (1990) Synchronous differentiation of Trypanosoma brucei from bloodstream to procyclic forms in vitro. Eur J Biochem 192:373–378
Zilka A, Garlapati S, Dahan E, Yaolsky V, Shapira M (2001) Developmental regulation of heat shock protein 83 in Leishmania. 3′ processing and mRNA stability control transcript abundance, and translation id directed by a determinant in the 3′-untranslated region. J Biol Chem 276:47922–47929
Zou J, Guo Y, Guettouche T, Smith DF, Voellmy R (1998) Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 complex) that forms a stress-sensitive complex with HSF1. Cell 94:471–480
Acknowledgements
The authors gratefully acknowledge the support of the Wellcome Trust.
Conflicts of interest
The authors declare that they have no conflicts of interest.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling Editor: David Robinson
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Schwede, A., Kramer, S. & Carrington, M. How do trypanosomes change gene expression in response to the environment?. Protoplasma 249, 223–238 (2012). https://doi.org/10.1007/s00709-011-0282-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00709-011-0282-5