Abstract
The retinoblastoma tumor suppressor protein (pRb) regulates cell cycle progression by controlling the G1-to-S phase transition. As evidenced in mammals, pRb has three functionally distinct binding domains and interacts with a number of proteins including the E2F family of transcription factors, proteins with a conserved LxCxE motif (D-type cyclin), and c-Abl tyrosine kinase. CDK-mediated phosphorylation of pRb inhibits its ability to bind target proteins, thus enabling further progression of the cell cycle. As yet, the roles of pRb and pRb-binding factors have not been well characterized in plants. By using antibody which specifically recognizes phosphorylated serines (S807/811) in the c-Abl tyrosine kinase binding C-domain of human pRb, we provide evidence for the cell cycle-dependent changes in pRb-like proteins in root meristems cells of Vicia faba. An increased phosphorylation of this protein has been found correlated with the G1-to-S phase transition.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
A vast number of similarities and differences have been shown to interlace in structure, function, and mutual relationships between the principal regulators of the cell division cycles in yeast, mammals, and plants (de Jager et al. 2005). The entry into S phase in mammalian cells, but not in yeasts, is controlled by key regulators of the retinoblastoma protein (pRb) pathway (Weinberg 1995). Recently, Rb-related genes and their products called retinoblastoma-related proteins (RBRs) have been discovered in various plant species (Ach et al. 1997; Grafi et al. 1996; Miskolczi et al. 2007; Vandepoele et al. 2002; Xie et al. 1996). In mammals, pRb is a tumor suppressor protein that prevents cells from progressing through the cell cycle and mitotic divisions. Oncogenic proteins, such as those produced by virus-infected cells, can bind and inactivate pRb, eventually leading to tumorogenesis. Accordingly, in a number of cancer types, pRb was found to be dysfunctional (Das et al. 2005; Funk et al. 1997; Ludlow and Skuse 1995; Nevins 2001; Sellers and Kaelin 1997). RBRs are also involved in DNA repair, DNA damage checkpoint control, differentiation, cellular senescence, and apoptosis.
pRb is a member of the pocket protein family which has three distinct binding domains: the first, which binds proteins with the LxCxE motif (e.g., D-type cyclins and oncoproteins; Dowdy et al. 1993; Zamanian and LaThangue 1992); the second, large A/B pocket, which binds the E2F family of transcription factors and the third, C (more divergent in plant RBRs), which binds the c-Abl tyrosine kinase in mammals (Knudsen and Wang 1996; Korenjak and Brehm 2005; Münger and Howley 2002).
The activity of pRb is regulated by phosphorylation, catalyzed by several cyclin-dependent protein kinases (CDKs). The type of phosphorylation distinctly affects the ability of pRb to interact with its various partner proteins. Accordingly, Thr 821/826 phosphorylation is required to inhibit pRb adjoining to LxCxE-containing proteins (Knudsen and Wang 1996), S780 phosphorylation blocks the ability of Rb to bind E2F (Kitagawa et al. 1996; Lundberg and Weinberg 1998), while Ser 807/811 phosphorylation disrupts c-Abl binding (Knudsen and Wang 1996).
Near the end of mitosis, phosphatase-mediated activation of pRb allows this protein to bind and to inhibit E2F family transcription factors connected with DP protein (Brown et al. 1999; Münger and Howley 2002). As long as the transcription activating E2F–DP complexes are blocked by pRb, the progression through the cell cycle is halted and the cell remains in the G1 phase (De Veylder et al. 2003; Funk et al. 1997; de Jager et al. 2005; Ludlow and Skuse 1995; Nevins 2001). In addition, pRb-E2F/DP complex recruits histone deacetylase (HDAC) to chromatin, exerting an inhibitory effect on DNA synthesis. When it is time for a cell to progress through the G1/S transition, CDK/cyclin complexes (CDK4 and CDK6 associated with cyclin D, and CDK2 with cyclin E in mammals) phosphorylate and inhibit pRb activity (Das et al. 2005; Korenjak and Brehm 2005; Münger and Howley 2002; Sellers and Kaelin 1997; Weinberg 1995). The initial CDK4,6/cyclin D-mediated phosphorylation is followed by phosphorylation performed by CDK2/cyclin E. All these processes lead to dissociation of pRb from E2F/DP, and consequently, to transcriptional activation of genes required for the onset of S phase, including proliferating cell nuclear antigen (PCNA), which enhances DNA replication by helping polymerase to attach to the DNA (Das et al. 2005; De Veylder et al. 2003; Funk et al. 1997).
Similarly, the tyrosine kinase activity of nuclear c-Abl (a product of the proto-oncogene c-abl) was found to be negatively regulated by a specific interaction of its C-terminal ATP-binding lobe with pRB. When pRb is phosphorylated on Ser 807/811 during S phase, c-Abl dissociates and becomes activated to bind DNA through an essential DNA-binding domain. This process allows c-Abl to participate directly in the regulation of transcription (Welch and Wang 1993). The cell cycle-dependent DNA binding activity of c-Abl is regulated by cdc2-mediated phosphorylation of the large C-terminal segment at three sites during interphase and at seven additional sites during mitosis, which abolishes association of c-Abl with chromatin. Mutant mice expressing tyrosine kinase lacking DNA-binding domain are lethal due to multiple defects at birth (Kipreos and Wang 1992).
Recent evidence has suggested that the control mechanisms leading to the G1/S transition are conserved between animals and plants. Plant cells possess several D-type cyclins (expressed during the G1 phase) that contain an LxCxE motif (Soni et al. 1995). The RBRs interact through LxCxE binding site with D-type cyclins and geminivirus replication proteins (Ach et al. 1997; Arguello-Astorga et al. 2004). The amount of plant pRb phosphorylated in a cell cycle-dependent manner by CDKA–cyclin D complexes (analogous to mammalian CDK4,6/CycD) reaches its maximum level at the G1–S transition (Boniotti and Gutierrez 2001; De Veylder et al. 2003; Gutierrez et al. 2002; Nakagami et al. 2002). Moreover, three E2Fs, two DPs, and three monomeric E2F-like proteins identified in plants reveal both a domain organization similar to their mammalian counterparts and bind the same DNA-sequence motifs regulating the expression of genes involved in DNA synthesis (Albani et al. 2000; Vandepoele et al. 2002). However, no data are available at present relevant to either c-Abl kinase homologs or phosphorylation sites in plant RBR proteins. By using antibodies recognizing the amino acid sequence surrounding local phosphorylation site (S807/811) of pRb C-domain responsible for binding c-Abl tyrosine kinase in human cells, we provide data suggesting that there may be functional link between posttranslational modifications of RBRs and successive events that drive the cell cycle progression in primary root meristem cells of Vicia faba.
Materials and methods
Immunocytochemistry
Roots of the 4-day-old seedlings of Vicia faba L. subsp. minor were fixed for 30 min in a freshly prepared 4% formaldehyde buffered with PBS (pH 7.4). Meristems were cut off, rinsed in PBS three times (5 min each) and treated with an enzyme solution (1% cellulose Onozuka R-10, 1% pectolyase Y-23', and 1% pectinase) in PBS for 40 min (pH 5.3). After washing with PBS, cells were attached to Super Frost Plus glass slides, and incubated for 60 min with PBS-buffered 5% BSA (w/v) solution containing 0.5% Triton X-100. After washing with PBS/0.5% Triton X-100 (PBT), the samples were processed for immunostaining. The antibody against phospho-Rb (Ser 807/811) produced by rabbits immunized with phosphopeptide corresponding to residues around Ser 807/811 of human Rb (Cell Signaling) were overlayed onto the cells (1:100). After incubation (overnight, at 4°C) the slides were washed three times (5 min each) with PBT and incubated in the secondary anti-rabbit fluorescein isothiocyanate (FITC)-conjugated antibody (Cell Signaling) at a dilution of 1:200 in PBT for 2 h at room temperature in dark. Then, the samples were washed two times with PBT and PBS (5 min each). Negative control sections were incubated with nonimmune rabbit serum in place of primary antibody. DNA was stained with 1 μg mL−1 DAPI (4,6-diamino-2-phenylindole dihydrochloride) in PBS. Samples were embedded in PBS/glycerol mixture (9:1) with 2.3% DABCO (Sigma-Aldrich).
Observations were made using Optiphot-2 fluorescence microscope (Nikon, Poland) equipped with B-2A filter (λ = 450–490 nm) for FITC and UV-2A filter (λ = 360–460 nm) for DAPI. For analyses of phosphoprotein levels and DNA contents, all images were recorded by DXM 1200 CCD (Nikon) camera at exactly the same time of integration. The data scored from 50 cells from each phase of the cell cycle served to calculate mean values (±SD) for each experimental series. Fluorescence signals were computed, and nuclei or cells plot profiles were done using Scion Image for Windows (Polit and Maszewski 2004).
Western blot analysis
The control and partially synchronized root meristem cells of V. faba (18 h treatment with 1.25 mM hydroxyurea—HU, followed by 4, 6, 8, 10 h recovery) were homogenized using 50 mM Tris HCl buffer, pH 8.0, containing 75 mM NaCl, 0.5 mM MgCl2, 20 mM EDTA, 1% Nonidet p-40, and Protease Inhibitor Cocktail (Sigma P-9599) added according to protocol. To check whether detected bands were phosphorylated, portion of root homogenate was treated with calf intestinal alkaline phosphatase (5 U, 20 min, Sigma). The samples were cleared by centrifugation, and total protein extracts were fractionated on 4–12% Bis Tris/MES PAGE-SDS gels (Novex) and blotted onto nitrocellulose membrane (0.45 μm pore size; Invitrogen). Proteins containing the amino acid sequence corresponding to the residues around phospho-Ser 807/811 of human Rb were detected with the rabbit polyclonal anti-phospho-Rb (Ser 807/811) antibodies (1:1,000 dilution; Cell Signaling). Secondary peroxidase-conjugated goat anti-rabbit antibodies (Abcam) were applied at a dilution of 1:2,000 and revealed with enhanced chemiluminescence detection reagents (Pierce).
Results and discussion
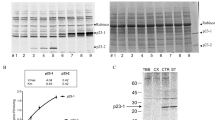
It becomes increasingly apparent that most of the cell cycle control mechanisms are similarly conserved in plants and animals (De Veylder et al. 2003; de Jager et al. 2005; Gutierrez et al. 2002). Considering the occurrence of RBRs in unicellular green algae (e.g., a single-copy Mat3 homolog of pRb in Chlamydomonas reinhardtii; Umen and Goodenough 2001), the presence of gender-specific RBR1 in Volvox carteri (Hallman 2009), and the role of RBRs in embryogenesis, differentiation, and cell fate determination in plants (Gordon-Kamm et al. 2002; Sabelli et al. 2005), our current work has aimed at recognizing possible connections between the posttranslational modifications of RBRs and cell division cycle progression in root meristem cells of V. faba. In immunoblots of all protein samples extracted either from the control root cells or after their hydroxyurea-mediated synchronization treatment, the antibody raised against human phospho-Rb (Ser 807/811) detected only one band at the position of about 72 kDa, but did not detect this band after alkaline phosphatase treatment (Fig. 1). Although the estimated molecular weight of RBR in V. faba was found lower than that of most pRb family members (e.g., Ábrahám et al. 2010), it corresponds well with the values presented for maize open reading frame capable of encoding a ZmRb1 protein consisting of 683 amino acids (Xie et al. 1996) and with the results of Taieb et al. (1998) on Xenopus laevis oocytes. In the control experimental series using nonimmune rabbit serum, no specific immunostaining could be observed (Fig. 2).
Immumoblotting analysis of pRb-like proteins in root meristems of V. faba using anti-phospho-Rb (S807/811) antibody; lysates of the control cells (a) and after partial synchronization with 1.25 mM HU, followed by 4 h recovery—S phase, 6 h recovery—G2 phase, 8 h recovery—G2/M/G1 phase, and 10 h recovery—G1/S phase (b). Lane I—molecular weight marker, lane II—SDS-Nu-PAGE electrophoresis and Coomasie staining, lanes III and IV—Western blots of separated and electrotransferred proteins performed using primary rabit anti-phospho-Rb (Ser 807/811) IgG fraction and secondary goat anti-rabbit IgG antibody conjugated with peroxidase, lane IV—cell lysate treated with calf intestinal alkaline phosphatase before electrophoresis
Despite nearly equal Western blot labeling detected in samples from all partially synchronized root cell populations, immunofluorescence of phosphorylated S807/811 residues in the 72 kDa RBR-like protein revealed evident cell cycle phase-dependent changes (Figs. 1, 3, and 4). The multiple regions of phosphorylation could already be seen in two groups of daughter chromosomes decondensing in late telophase during early stages of cell wall formation (Fig. 3, P and Q). Accordingly, at the onset of a new cell division cycle, each nucleus contains same amount of phosphorylated RBR-like protein (Fig. 3, A). Both direct observations and quantitative measurements of nuclear fluorescence indicate that the intensity of immunolabeling increases during the G1/S transition (Fig. 3, A–C; Fig. 4), then gradually diminishes towards prophase (Fig. 3, D–I) and disappears almost completely at later stages of mitosis (i.e., during metaphase, anaphase, and early telophase; Fig. 3, K–N; Fig. 4). It seems probable, then, that an overlap between the time patterns of RBR phosphorylation in V. faba and pRb phosphorylation and mammals represents a common regulatory pathway, by which the release and activation of c-Abl kinase is followed by phosphorylation of tyrosine residues on the carboxy-terminal repeated domain of the RNA polymerase II. Consequently, such process would implicate that the conserved mechanism operates in plants to coordinate multiple nuclear components indispensable for transcriptional activation of genes engaged in the progression of DNA replication (comp. Baskaran et al. 1993, 1997; Duyster et al. 1995; Kipreos and Wang 1992; Pendergast 1996; Welch and Wang 1993).
Mean intensity of fluorescence (MIF) of proteins detected by anti-phospho-Rb (S807/811)-FITC in the double-stained (FITC/DAPI) root meristem cells of V. faba during successive phases of the cell cycle; G1, S, G2, P prophase, M metaphase, A anaphase, T telophase, PT post-telophase. Pixel value = MIF/pel ± SD
Although, during interphase, most of the nucleoli have been found devoid of the phosphorylated form of RBR-like protein (Fig. 3, A, B, and D; Fig. 4), a small fraction of late S/early G2 cells (selected by DAPI-DNA cytofluorometry and cell size measurements) revealed an intense immunolabeling, considerably exceeding that observed in the extranucleolar nucleoplasm (Fig. 3, E). The appearance of such cells may indicate that a large amount of RBR-like molecules phosphorylated on S807/811 has to be localized into the nucleoli within a relatively short period of time corresponding to the transition towards the premitotic stage of interphase. Since no analogy of this phenomenon can be drawn to animals, it seems reasonable to assume that it may contribute to some additional plant-specific regulatory mechanisms controlling nuclear metabolism, the activity of RNA I polymerase, or cell cycle progression.
Evident variation in the intranuclear distribution of phospho-RBR-like protein revealed by direct microscopic observation led us to correlate the intensities of immunolabeling (FITC) and DAPI-DNA staining by using fluorescence profiling. Corresponding linescans obtained from selected cell nuclei (Fig. 5a–d) clearly indicate an inverse relationship between the concentration of nuclear DNA (density of chromatin) and the strength of the signal emitted from FITC-conjugated antibody. Apart from an intense immunostaining detected in nuclear areas characterized by a relatively low level of chromatin condensation (Fig. 5a, b), a significant amount of FITC-fluorescence could be observed within the perinucleolar regions, usually occupied by condensed chromatin. However, the linescan profiles across the boundary of nucleoli and nucleoplasm derived from DAPI stained early S-phase nuclei revealed a reduced degree of chromatin compaction, which may correlate with phospho-RBR-mediated DNA replication rather, than with a suppressed metabolic activity of the perinucleolar compartment (Fig. 5c, d).
When compared with the areas of phospho-RBR-like protein observed in the G1 or S phase nuclei, the immunopositive localities in the G2 nuclei could be seen as loosely packed fluorescing regions scattered over the dark chromatin background. At prophase, they were still discernible in the form of larger but less numerous clusters between the condensing chromosomes. After nuclear envelope breakdown, during prometaphase, remnants of phosphorylated RBR-like protein migrated into the perinuclear cytoplasm. The phospho-positive cytoplasmic inclusions were also visible as homogenous bright areas in the vicinity of metaphase chromosomes and as bright strands spread along the arms of some chromosomes during early anaphase. At later stages of anaphase and during early telophase, immunonegative areas corresponded to chromosome territories separated by a slightly brighter homogenous area of cytoplasm in the central part of a cell.
The above data are in agreement with the observations of Szekely et al. (1991) and their results concerning subcellular localization of pRb in primate cell lines. Using immunofluorescence staining with different monoclonal and polyclonal antibodies pRb granules, heterogeneous in size, were localized over the interphase nuclei predominantly in euchromatic areas with low DNA density. The largest Rb positive regions lined up on the heterochromatin/euchromatin boundary. During mitosis, the Rb protein dissociated from the condensing chromosomes, then dispersed throughout the cytoplasm during metaphase and anaphase, and associated again with the decondensing chromatin at telophase.
Conclusions
Our present data indicate that plant cells contain the RBR-like protein recognized by the anti-human phospho-Rb antibody. Similarly to mammalian cells, serine residues (S807/811) located in the carboxyl-terminal regulatory domain of this protein undergo phosphorylation suggesting the occurrence of a nuclear factor capable both to recognize this motif and to interact with other components of chromatin in a cell cycle-dependent manner. However, considering a relatively low molecular mass of RBR-like protein in V. faba, one cannot exclude that it has been submitted to elaborate evolutionary modifications, as it is the case with other elements of the cell cycle machinery in plants.
References
Ach RA, Durfee T, Miller AB, Taranto P, Hanley-Bowdoin L, Zambryski PC, Gruissem W (1997) RRB1 and RRB2 encode maize retinoblastoma-related proteins that interact with a plant D-type cyclin and geminivirus replication protein. Mol Cell Biol 17:5077–5086
Albani D, Mariconti L, Ricagno S, Pitto L, Moroni C, Helin K, Cella R (2000) DcE2F, a functional plant E2-like transcriptional activator from Daucus carota. J Biol Chem 275:19258–19267
Arguello-Astorga G, Lopez-Ochoa L, Kong L-J, Orozco BM, Settlage SB, Hanley-Bowdoin L (2004) A novel motif in geminivirus replication proteins interacts with the plant retinoblastoma-related protein. J Virol 78:4817–4826
Ábrahám E, Miskolczi P, Ayadin F, Yu P, Kotogány E, Bakó L, Ötvös K, Horváth GV, Dudits D (2010) Immunodetection of retinoblastoma-related protein and its phosphorylated form in interphase and mitotic alfalfa cells. J Exp Bot. doi:10.1093/jxb/erq413
Baskaran R, Chiang GG, Mysliwiec T, Kruh GD, Wang JYJ (1997) Tyrosine phosphorylation of RNA polymerase II carboxyl-terminal domain by the Abl-related gene product. J Biol Chem 272:18905–18909
Baskaran R, Dahmus ME, Wang JYJ (1993) Tyrosine phosphorylation of mammalian RNA polymerase II carboxyl-terminal domain. Proc Natl Acad Sci USA 90:11167–11171
Boniotti MB, Gutierrez C (2001) A cell-cycle-regulated kinase activity phosphorylates plant retinoblastoma protein and contains, in Arabidopsis, a CDKA/cyclin D complex. Plant J 28:341–350
Brown VD, Phillips RA, Gallie BL (1999) Cumulative effect of phosphorylation of pRB on regulation of E2F activity. Mol Cell Biol 19:3246–3256
Das SK, Hashimoto T, Shimizu K, Yoshida T, Sakai T, Sowa Y, Komoto A, Kanazawa K (2005) Fucoxanthin induces cell cycle arrest at G0/G1 phase in human colon carcinoma cells through up-regulation of p21WAF1/Cip1. Biochim Biophys Acta 1726:328–335
De Veylder L, Joubes J, Inzé D (2003) Plant cell cycle transitions. Curr Opin Plant Biol 6:536–543
Dowdy SF, Hinds PW, Louie K, Reed SI, Arnold A, Weinberg RA (1993) Physical interaction of the retinoblastoma protein with human D cyclins. Cell 73:499–511
Duyster J, Baskaran R, Wang JYJ (1995) Src homology 2 domain as a specificity determinant in the c-Abl-mediated tyrosine phosphorylation of the RNA polymerase II carboxyl-terminal repeated domain. Proc Natl Acad Sci USA 92:1555–1559
Funk JO, Waga S, Harry JB, Espling E, Stillman B, Galloway DA (1997) Inhibition of CDK activity and PCNA-dependent DNA replication by p21 is blocked by interaction with the HPV-16 E7 oncoprotein. Genes Dev 11:2090–2100
Gordon-Kamm W, Dilkes BP, Lowe K, Hoerster G, Sun X, Ross M, Church L, Bunde C, Farrell J, Hill P, Maddock J, Sykes L, Li Z, Woo Y, Bidney D, Larkins BA (2002) Stimulation of the cell cycle and maize transformation by disruption of the plant retinoblastoma pathway. Proc Natl Acad Sci USA 99:11975–11980
Grafi G, Burnett RJ, Helentjaris T, Larkins BA, DeCaprio JA, Sellers WR, Kaelin WG Jr (1996) A maize cDNA encoding a member of the retinoblastoma protein family: involvement in endoredulication. Proc Natl Acad Sci USA 93:8962–8967
Gutierrez C, Ramirez-Parra E, Castellano MM, del Pozo JC (2002) G1 to S transition: more than a cell cycle engine switch. Curr Opin Plant Biol 5:480–486
Hallman A (2009) Retinoblastoma-related proteins in lower eukaryotes. Commun Integr Biol 2:538–544
de Jager SM, Maughan S, Dewitte W, Scofield S, Murray JAH (2005) The developmental context of cell-cycle control in plants. Semin Cell Dev Biol 16:385–396
Kipreos ET, Wang JY (1992) Cell cycle-regulated binding of c-Abl tyrosine kinase to DNA. Science 256:382–385
Kitagawa M, Higashi H, Jung HK, Suzuki-Takahashi I, Ikeda M, Tamai K, Kato J, Segawa K, Yoshida E, Nishimura S, Taya Y (1996) The consensus motif for phosphorylation by cyclin D1–Cdk4 is different from that for phosphorylation by cyclin A/E–Cdk2. EMBO J 15:7060–7069
Knudsen ES, Wang JY (1996) Differential regulation of retinoblastoma protein function by specific Cdk phosphorylation sites. J Biol Chem 271:8313–8320
Korenjak M, Brehm A (2005) E2F–Rb complexes regulating transcription of genes important for differentiation and development. Curr Opin Genet Dev 15:520–527
Ludlow JW, Skuse GR (1995) Viral oncoprotein binding to pRB, p107, p130 and p300. Virus Res 35:113–121
Lundberg AS, Weinberg RA (1998) Functional inactivation of the retinoblastoma protein requires sequential modification by at least two distinct cyclin-cdk complexes. Mol Cell Biol 18:753–761
Miskolczi P, Lendvai Á, Horváth GV, Pettkó-Szandtner A, Dudits D (2007) Conserved functions of retinoblastoma proteins: from purple retina to green plant cells. Plant Sci 172:671–683
Münger K, Howley PM (2002) Human papillomavirus immortalization and transformation functions. Virus Res 89:213–228
Nakagami H, Kawamura K, Sugisaka K, Sekine M, Shinmyo A (2002) Phosphorylation of retinoblastoma-related protein by the cyclin D/cyklin-dependent kinase complex is activated at the G1/S-phase transition in tobacco. Plant Cell 14:847–1857
Nevins JR (2001) The Rb/E2F pathway and cancer. Hum Mol Genet 10:699–703
Pendergast AM (1996) Nuclear tyrosine kinases: from Abl to WEE1. Curr Opin Cell Biol 8:174–181
Polit J, Maszewski J (2004) Protein phosphorylation during the transitions from G1 arrest point to S and G2 arrest point to M in Vicia faba root meristem. Biol Plant 48:351–359
Sabelli PA, Dante RA, Leiva-Neto JT, Jung R, Gordon-Kamm WJ, Larkins BA (2005) RBR3, a member of the retinoblastoma-related family from maize, is regulated by the RBR1/E2F pathway. Proc Natl Acad Sci USA 102:13005–13012
Sellers WR, Kaelin WG Jr (1997) Role of the retinoblastoma protein in the pathogenesis of human cancer. J Clin Oncol 15:3301–3312
Soni R, Carmichael JP, Shah ZH, Murray JAH (1995) A family of cyclin D homologs from plants differentially controlled by growth regulators and containing the conserved retinoblastoma protein interaction motif. Plant Cell 7:85–103
Szekely L, Uzvolgyi E, Jiang WQ, Durko M, Wiman KG, Klein G, Sumegi J (1991) Subcellular localization of the retinoblastoma protein. Cell Growth Differ 2:287–295
Taieb F, Karaiskou A, Rime H, Jessus C (1998) Human retinoblastoma protein (Rb) is phosphorylated by cdc2 kinase and MAP kinase in Xenopus maturing oocytes. FEBS Lett 425:465–471
Umen JG, Goodenough UW (2001) Control of cell division by a retinoblastoma protein homolog in Chlamydomonas. Genes Dev 15:1652–1661
Vandepoele K, Raes J, De Veylder L, Rouzé P, Rombauts S, Inzé D (2002) Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 14:903–916
Weinberg RA (1995) The retinoblastoma protein and cell cycle control. Cell 81:323–330
Welch PJ, Wang JY (1993) A C-terminal protein-binding domain in the retinoblastoma protein regulates nuclear c-Abl tyrosine kinase in the cell cycle. Cell 75:779–790
Xie Q, Sanz-Burgos AP, Hannon GJ, Gutiérrez C (1996) Plant cells contain a novel member of the retinoblastoma family of growth regulatory proteins. EMBO J 15:4900–4908
Zamanian M, LaThangue NB (1992) Adenovirus E1a prevents the retinoblastoma gene product from repressing the activity of a cellular transcription factor. EMBO J 11:2603–2610
Acknowledgments
We thank Prof. Janusz Maszewski from the Department of Cytophysiology, University of Łódź, for his consultation and discussion as well as for his help in preparing the English version of the manuscript. This work was partially supported by grant from the University of Łódź, No. 505/0391.
Conflict of interest
The authors declare that they have no conflict of interest.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling Editor: Heiti Paves
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Polit, J.T., Kaźmierczak, A. & Walczak-Drzewiecka, A. Cell cycle-dependent phosphorylation of pRb-like protein in root meristem cells of Vicia faba . Protoplasma 249, 131–137 (2012). https://doi.org/10.1007/s00709-011-0272-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00709-011-0272-7