Abstract
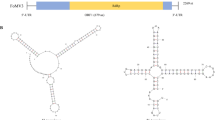
In this study, we determined the complete genome sequence of a novel totivirus, tentatively named "Mangifera indica totivirus 1" (MiTV1), identified in ‘Apple’ mango in China. The double-stranded RNA genome of MiTV1 is 4800 base pairs (bp) in length and contains two open reading frames (ORFs) encoding a putative coat protein (CP) and an RNA-dependent RNA polymerase (RdRp). Phylogenetic analysis based on RdRp and CP amino acid sequences showed that MiTV1 is closely related to members of the genus Totivirus in the family Totiviridae. To our knowledge, this is the first report of a totivirus found in Mangifera indica.


Similar content being viewed by others
Data availability
The complete genome sequence reported here is available in the GenBank database under the accession number OR860451.
References
Akinyemi IA, Wang F, Chang ZX, Wu Q (2018) Genome characterization of the newly identified maize-associated totivirus Anhui. Arch Virol 163(10):2929–2931
Alvarez-Quinto RA, Espinoza-Lozano RF, Mora-Pinargote CA, Quito-Avila DF (2017) Complete genome sequence of a variant of maize-associated totivirus from Ecuador. Arch Virol 162(4):1083–1087
Athoo TO, Winkler A, Knoche M (2020) Russeting in ‘Apple’ Mango: Triggers and Mechanisms. Plants (Basel) 9(7):898
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120
Buchfink B, Xie C, Huson DH (2015) Fast and sensitive protein alignment using DIAMOND. Nat Methods 12(1):59–60
Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T (2009) trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25(15):1972–1973
Chen S, Cao L, Huang Q, Qian Y, Zhou X (2016) The complete genome sequence of a novel maize-associated totivirus. Arch Virol 161(2):487–490
Cornejo-Franco JF, Alvarez-Quinto RA, Mollov D, Quito-Avila DF (2023) Identification and genetic characterization of a new totivirus from Bursera graveolens in western Ecuador. Arch Virol 168(4):102
Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, MacManes MD, Ott M, Orvis J, Pochet N, Strozzi F, Weeks N, Westerman R, William T, Dewey CN, Henschel R, LeDuc RD, Friedman N, Regev A (2013) De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc 8(8):1494–1512
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30(4):772–780
Liu X, Gorovsky MA (1993) Mapping the 5’ and 3’ ends of Tetrahymena thermophila mRNAs using RNA ligase mediated amplification of cDNA ends (RLM-RACE). Nucleic Acids Res 21(21):4954–4960
Morris TJ, Dodds JA (1979) Isolation and analysis of double-stranded RNA from virus-infected plant and fungal tissue. Phytopathoglogy 69(8):854–858
Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32(1):268–274
Read DA, Featherston J, Rees DJG, Thompson GD, Roberts R, Flett BC, Mashingaidze K, Pietersen G, Kiula B, Kullaya A, Mbega ER (2019) Diversity and distribution of Maize-associated totivirus strains from Tanzania. Virus Genes 55(3):429–432
Sela N, Luria N, Yaari M, Prusky D, Dombrovsky A (2016) Genome sequence of a potential New Benyvirus isolated from Mango RNA-seq data. Genome Announc 4(6):e01250–e01216
Tan H, Zhao R, Wang H, Huang X (2023) Identification and molecular characterization of a novel member of the genus Totivirus from Areca catechu L. Arch Virol 168(10):247
Umer M, Liu J, You H, Xu C, Dong K, Luo N, Kong L, Li X, Hong N, Wang G, Fan X, Kotta-Loizou I, Xu W (2019) Genomic, Morphological and Biological Traits of the Viruses Infecting Major Fruit Trees. Viruses 11(6):515
Walker PJ, Siddell SG, Lefkowitz EJ, Mushegian AR, Adriaenssens EM, Alfenas-Zerbini P, Dempsey DM, Dutilh BE, Garcia ML, Curtis Hendrickson R, Junglen S, Krupovic M, Kuhn JH, Lambert AJ, Lobocka M, Oksanen HM, Orton RJ, Robertson DL, Rubino L, Sabanadzovic S, Simmonds P, Smith DB, Suzuki N, Van Doorslaer K, Vandamme AM, Varsani A, Zerbini FM (2022) Recent changes to virus taxonomy ratified by the International Committee on Taxonomy of Viruses (2022). Arch Virol 167(11):2429–2440
Warschefsky EJ, von Wettberg EJB (2019) Population genomic analysis of mango (Mangifera indica) suggests a complex history of domestication. New Phytol 222(4):2023–2037
Xie J, Wei D, Jiang D, Fu Y, Li G, Ghabrial S, Peng Y (2006) Characterization of debilitation-associated mycovirus infecting the plant-pathogenic fungus Sclerotinia sclerotiorum. J Gen Virol 87(Pt 1):241–249
Acknowledgements
The authors thank Quansheng Yao, Songbiao Wang, and Xiaowei Ma for assistance with the lab and mango resource.
Funding
This work was financially supported by the Central Public-interest Scientific Institution Basal Research Fund (NO. 1630062023006).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
This article does not contain any studies involving human participants or animals.
Conflict of interest
The authors declare have no competing interests.
Additional information
Communicated by Robert H.A. Coutts
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chen, M., Xia, Y. & Wang, Q. Identification and molecular characterization of a novel totivirus from Mangifera indica. Arch Virol 169, 58 (2024). https://doi.org/10.1007/s00705-024-06001-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00705-024-06001-x