Abstract
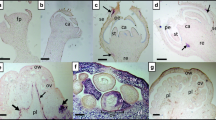
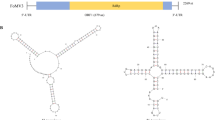
A novel emaravirus, tentatively named “clematis yellow mottle associated virus” (CYMaV), was identified through transcriptome sequencing and RT-PCR analysis of yellow-mottled leaf samples from Clematis brevicaudata DC. The genome of CYMaV consists of five viral RNAs: RNA1 (6591 nucleotides, nt), RNA2 (1982 nt), RNA3a (1301 nt), RNA3b (1397 nt), and RNA4 (1192 nt). The 13-nt sequences at the 5′- and 3′-termini of the CYMaV RNAs are conserved and have reverse complementary, as typically seen in emaraviruses. The proteins encoded by CYMaV shared the highest amino acid sequence similarity with those of the unclassified Karaka Okahu purepure emaravirus (KOPV), with 60.2% identity in the RNA-dependent RNA polymerase (RdRp), 44.4% in the glycoprotein precursor, and 46.9% in the nucleocapsid protein. A phylogenetic tree based on amino acid sequences of the RdRp revealed that CYMaV is most closely related to KOPV and clusters with ChMaV (chrysanthemum mosaic-associated virus, LC576445) and PCLSaV (pear chlorotic leaf spot-associated virus, MK602177) in one distinct clade. Transmission electron microscopy observation of negatively stained samples from C. brevicaudata revealed spherical virus-like particles (VLPs) approximately 100 nm in diameter. Five primers, specific for each viral RNA, were used to detect CYMaV in 11 symptomatic and two asymptomatic C. brevicaudata samples, but the results failed to show a consistent association of viral infection with symptoms. CYMaV can be considered a putative new member in the genus Emaravirus, and this marks the first report of an emaravirus found infecting C. brevicaudata plants.


Similar content being viewed by others
References
Wang JY, Qiao Q, Zhang J, Hu FR (2020) Research progress and landscape application of Clematis L. J Shandong Agric Univ (Nat Sci) 51(02):217–221
Ma T, Xiao P, Nie L (2012) Research on the effective parts from Clematis brevicaudata DC. Chin J Ethnomed Ethnopharm 18(12):27–29
Sun HF, Wang H, Yang H, Li N, Wei MY, Yan Y (2022) First report of leaf spot on Clematis brevicaudata DC. caused by Alternaria alternata in China. Plant Dis. https://doi.org/10.1094/PDIS-04-22-0902-PDN
Elbeaino T, Digiaro M, Mielke-Ehret N, Muehlbach HP, Martelli, GP ICTV Report Consortium (2018) ICTV virus taxonomy profile: fimoviridae. J Gen Virol 99(11):1478–1479
Ilyas M, Avelar S, Schuch UK, Brown JK (2018) First Report of an Emaravirus Associated with Witches’ Broom Disease and Eriophyid Mite Infestations of the Blue Palo Verde Tree in Arizona. Plant Dis 102(9):1863
Kubota K, Usugi T, Tomitaka Y, Shimomoto Y, Takeuchi S, Kadono F, Yanagisawa H, Chiaki Y, Tsuda S (2020) Perilla mosaic virus is a highly divergent emaravirus transmitted by Shevtchenkella sp. (Acari: Eriophyidae). Phytopathology 110(7):1352–1361
Gupta AK, Hein GL, Tatineni S (2019) P7 and P8 proteins of High Plains wheat mosaic virus, a negative-strand RNA virus, employ distinct mechanisms of RNA silencing suppression. Virology 535:20–31
Elbeaino T, Digiaro M, Martelli GP (2009) Complete nucleotide sequence of four RNA segments of fig mosaic virus. Arch Virol 154(11):1719–1727
Rehanek M, von Bargen S, Bandte M, Karlin DG, Büttner C (2021) A novel emaravirus comprising five RNA segments is associated with ringspot disease in oak. Arch Virol 166(3):987–990
Kubota K, Yanagisawa H, Chiaki Y, Yamasaki J, Horikawa H, Tsunekawa K, Morita Y, Kadono F (2021) Complete nucleotide sequence of chrysanthemum mosaic-associated virus, a novel emaravirus infecting chrysanthemum. Arch Virol 166(4):1241–1245
Tatineni S, McMechan AJ, Wosula EN, Wegulo SN, Graybosch RA, French R, Hein GL (2014) An eriophyid mite-transmitted plant virus contains eight genomic RNA segments with unusual heterogeneity in the nucleocapsid protein. J Virol 88(20):11834–11845
Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, MacManes MD, Ott M, Orvis J, Pochet N, Strozzi F, Weeks N, Westerman R, William T, Dewey CN, Henschel R, LeDuc RD, Friedman N, Regev A (2013) De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc 8(8):1494–1512
Rabbidge LO, Blouin AG, Chooi KM, Higgins CM, MacDiarmid RM (2021) Characterisation and Distribution of Karaka Ōkahu Purepure Virus-A Novel Emaravirus Likely to Be Endemic to New Zealand. Viruses 13(8):1611
Amroun A, Priet S, de Lamballerie X, Quérat G (2017) Bunyaviridae RdRps: structure, motifs, and RNA synthesis machinery. Crit Rev Microbiol 43(6):753–778
Buzkan N, Chiumenti M, Massart S, Sarpkaya K, Karadağ S, Minafra A (2019) A new Emaravirus discovered in Pistacia from Turkey. Virus Res 263(2019)159–163
Rehanek M, Karlin DG, Bandte M, Al Kubrusli R, Nourinejhad Zarghani S, Candresse T, Büttner C, von Bargen S (2022) The Complex World of Emaraviruses-Challenges. Insights and Prospects Forests 13(11):1868
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61(3):539–542
Elbeaino T, Digiaro M, Alabdullah A, De Stradis A, Minafra A, Mielke N, Castellano MA, Martelli GP (2009) A multipartite single-stranded negative-sense RNA virus is the putative agent of fig mosaic disease. J Gen Virol 90(Pt 5):1281–1288
Acknowledgments
This work was supported by funds from the Natural Science Foundation of Liaoning Province (2021-MS-341) and the Program for Young and Middle-aged Scientific and Technological Innovation Talents of Shenyang City (RC210161).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
This article does not contain any experiments involving humans or animals performed by any of the authors.
Conflict of interest
The authors declare no competing interests.
Additional information
Communicated by Massimo Turina
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Below is the link to the electronic supplementary material
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yang, C., An, W., Li, C. et al. Detection and characterization of a putative emaravirus infecting Clematis brevicaudata DC. in China. Arch Virol 169, 10 (2024). https://doi.org/10.1007/s00705-023-05945-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00705-023-05945-w