Abstract
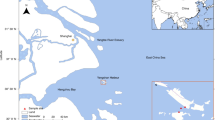
RNA viruses in marine environments have long been recognized as major players in the virosphere. In this study, the complete genome sequence of an RNA virus from Yangshan Harbor, named marine RNA virus Yangshan-LWW (YS-LWW), was obtained based on metavirome assembly. The genome of YS-LWW is 8839 nt in length and contains two open reading frames (ORFs). Both RdRP- and whole-genome-based phylogenetic analysis showed that YS-LWW, together with 45 viral isolates with sequences in public datasets, represents a new species in the genus Locarnavirus of the family Marnaviridae. PCR and public dataset mining indicate that YS-LWW and YS-LWW-like viruses have been widely detected in coastal and freshwater environments, suggesting that they might play a significant role in aquatic ecosystems.






Similar content being viewed by others
References
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL (2009) BLAST+: architecture and applications. BMC Bioinformatics 10:421
Camargo AP, Nayfach S, Chen IA, Palaniappan K, Ratner A, Chu K, Ritter SJ, Reddy TBK, Mukherjee S, Schulz F, Call L, Neches RY, Woyke T, Ivanova NN, Eloe-Fadrosh EA, Kyrpides NC, Roux S (2023) IMG/VR v4: an expanded database of uncultivated virus genomes within a framework of extensive functional, taxonomic, and ecological metadata. Nucleic Acids Res 51:D733–D743
Charon J, Kahlke T, Larsson ME, Abbriano R, Commault A, Burke J, Ralph P, Holmes EC (2022) Diverse RNA Viruses Associated with Diatom, Eustigmatophyte, Dinoflagellate, and Rhodophyte Microalgae Cultures. J Virol 96:e0078322
Chase EE, Monteil-Bouchard S, Gobet A, Andrianjakarivony FH, Desnues C, Blanc G (2021) A High Rate Algal Pond Hosting a Dynamic Community of RNA Viruses. Viruses 13
Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R (2020) TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol Plant 13:1194–1202
Chen YM, Sadiq S, Tian JH, Chen X, Lin XD, Shen JJ, Chen H, Hao ZY, Wille M, Zhou ZC, Wu J, Li F, Wang HW, Yang WD, Xu QY, Wang W, Gao WH, Holmes EC, Zhang YZ (2022) RNA viromes from terrestrial sites across China expand environmental viral diversity. Nat Microbiol 7:1312–1323
Culley AI, Lang AS, Suttle CA (2007) The complete genomes of three viruses assembled from shotgun libraries of marine RNA virus communities. Virol J 4:69
Darzentas N (2010) Circoletto: visualizing sequence similarity with Circos. Bioinformatics 26:2620–2621
Edgar RC, Taylor J, Lin V, Altman T, Barbera P, Meleshko D, Lohr D, Novakovsky G, Buchfink B, Al-Shayeb B, Banfield JF, de la Pena M, Korobeynikov A, Chikhi R, Babaian A (2022) Petabase-scale sequence alignment catalyses viral discovery. Nature 602:142–147
Greninger AL, DeRisi JL (2015) Draft Genome Sequences of Marine RNA Viruses SF-1, SF-2, and SF-3 Recovered from San Francisco Wastewater. Genome Announc 3
Hoff KJ, Stanke M (2013) WebAUGUSTUS–a web service for training AUGUSTUS and predicting genes in eukaryotes. Nucleic Acids Res 41:W123–128
Hufsky F, Beslic D, Boeckaerts D, Duchene S, Gonzalez-Tortuero E, Gruber AJ, Guo J, Jansen D, Juma J, Kongkitimanon K, Luque A, Ritsch M, Lencioni Lovate G, Nishimura L, Pas C, Domingo E, Hodcroft E, Lemey P, Sullivan MB, Weber F, Gonzalez-Candelas F, Krautwurst S, Perez-Cataluna A, Randazzo W, Sanchez G, Marz M (2022) The International Virus Bioinformatics Meeting 2022. Viruses 14
Hurwitz BL, Deng L, Poulos BT, Sullivan MB (2013) Evaluation of methods to concentrate and purify ocean virus communities through comparative, replicated metagenomics. Environ Microbiol 15:1428–1440
Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ (2010) Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119
Jiang Z, Liu J, Chen J, Chen Q, Yan X, Xuan J, Zeng J (2014) Responses of summer phytoplankton community to drastic environmental changes in the Changjiang (Yangtze River) estuary during the past 50 years. Water Res 54:1–11
Kolundzija S, Cheng DQ, Lauro FM (2022) RNA Viruses in Aquatic Ecosystems through the Lens of Ecological Genomics and Transcriptomics. Viruses 14
Lang AS, Vlok M, Culley AI, Suttle CA, Takao Y, Tomaru Y, Ictv Report C (2021) ICTV Virus Taxonomy Profile: Marnaviridae 2021. J Gen Virol 102
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359
Larsson A (2014) AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 30:3276–3278
Letunic I, Bork P (2021) Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res 49:W293–W296
Lu S, Wang J, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Marchler GH, Song JS, Thanki N, Yamashita RA, Yang M, Zhang D, Zheng C, Lanczycki CJ, Marchler-Bauer A (2020) CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res 48:D265–D268
Madeira F, Pearce M, Tivey ARN, Basutkar P, Lee J, Edbali O, Madhusoodanan N, Kolesnikov A, Lopez R (2022) Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res 50:W276–279
Nagasaki K (2008) Dinoflagellates, diatoms, and their viruses. J Microbiol 46:235–243
Nayfach S, Camargo AP, Schulz F, Eloe-Fadrosh E, Roux S, Kyrpides NC (2021) CheckV assesses the quality and completeness of metagenome-assembled viral genomes. Nat Biotechnol 39:578–585
Neri U, Wolf YI, Roux S, Camargo AP, Lee B, Kazlauskas D, Chen IM, Ivanova N, Zeigler Allen L, Paez-Espino D, Bryant DA, Bhaya D, Consortium RNAVD, Krupovic M, Dolja VV, Kyrpides NC, Koonin EV, Gophna U (2022) Expansion of the global RNA virome reveals diverse clades of bacteriophages. Cell 185:4023–4037 e4018
Nishimura Y, Yoshida T, Kuronishi M, Uehara H, Ogata H, Goto S (2017) ViPTree: the viral proteomic tree server. Bioinformatics 33:2379–2380
Rho M, Tang H, Ye Y (2010) FragGeneScan: predicting genes in short and error-prone reads. Nucleic Acids Res 38:e191
Roberts JMK, Anderson DL, Durr PA (2018) Metagenomic analysis of Varroa-free Australian honey bees (Apis mellifera) shows a diverse Picornavirales virome. J Gen Virol 99:818–826
Sadeghi M, Tomaru Y, Ahola T (2021) RNA Viruses in Aquatic Unicellular Eukaryotes. Viruses 13
Sayers EW, Beck J, Bolton EE, Bourexis D, Brister JR, Canese K, Comeau DC, Funk K, Kim S, Klimke W, Marchler-Bauer A, Landrum M, Lathrop S, Lu Z, Madden TL, O'Leary N, Phan L, Rangwala SH, Schneider VA, Skripchenko Y, Wang J, Ye J, Trawick BW, Pruitt KD, Sherry ST (2021) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 49:D10–D17
Sayers EW, O'Sullivan C, Karsch-Mizrachi I (2022) Using GenBank and SRA. Methods Mol Biol 2443:1–25
Sievers F, Higgins DG (2018) Clustal Omega for making accurate alignments of many protein sequences. Protein Sci 27:135–145
Simmonds P, Adriaenssens EM, Zerbini FM, Abrescia NGA, Aiewsakun P, Alfenas-Zerbini P, Bao Y, Barylski J, Drosten C, Duffy S, Duprex WP, Dutilh BE, Elena SF, Garcia ML, Junglen S, Katzourakis A, Koonin EV, Krupovic M, Kuhn JH, Lambert AJ, Lefkowitz EJ, Lobocka M, Lood C, Mahony J, Meier-Kolthoff JP, Mushegian AR, Oksanen HM, Poranen MM, Reyes-Munoz A, Robertson DL, Roux S, Rubino L, Sabanadzovic S, Siddell S, Skern T, Smith DB, Sullivan MB, Suzuki N, Turner D, Van Doorslaer K, Vandamme AM, Varsani A, Vasilakis N (2023) Four principles to establish a universal virus taxonomy. PLoS Biol 21:e3001922
Steward GF, Culley AI, Mueller JA, Wood-Charlson EM, Belcaid M, Poisson G (2013) Are we missing half of the viruses in the ocean? ISME J 7:672–679
Sun G, Xiao J, Wang H, Gong C, Pan Y, Yan S, Wang Y (2014) Efficient purification and concentration of viruses from a large body of high turbidity seawater. MethodsX 1:197–206
Tamura K, Stecher G, Kumar S (2021) MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol Biol Evol 38:3022–3027
UniProt C (2023) UniProt: the Universal Protein Knowledgebase in 2023. Nucleic Acids Res 51:D523–D531
Vlok M, Lang AS, Suttle CA (2019) Application of a sequence-based taxonomic classification method to uncultured and unclassified marine single-stranded RNA viruses in the order Picornavirales. Virus Evol 5:vez056
Wolf YI, Silas S, Wang Y, Wu S, Bocek M, Kazlauskas D, Krupovic M, Fire A, Dolja VV, Koonin EV (2020) Doubling of the known set of RNA viruses by metagenomic analysis of an aquatic virome. Nat Microbiol 5:1262–1270
Zayed AA, Wainaina JM, Dominguez-Huerta G, Pelletier E, Guo J, Mohssen M, Tian F, Pratama AA, Bolduc B, Zablocki O, Cronin D, Solden L, Delage E, Alberti A, Aury JM, Carradec Q, da Silva C, Labadie K, Poulain J, Ruscheweyh HJ, Salazar G, Shatoff E, Tara Oceans Coordinatorsdouble d, Bundschuh R, Fredrick K, Kubatko LS, Chaffron S, Culley AI, Sunagawa S, Kuhn JH, Wincker P, Sullivan MB, Acinas SG, Babin M, Bork P, Boss E, Bowler C, Cochrane G, de Vargas C, Gorsky G, Guidi L, Grimsley N, Hingamp P, Iudicone D, Jaillon O, Kandels S, Karp-Boss L, Karsenti E, Not F, Ogata H, Poulton N, Pesant S, Sardet C, Speich S, Stemmann L, Sullivan MB, Sungawa S, Wincker P (2022) Cryptic and abundant marine viruses at the evolutionary origins of Earth’s RNA virome. Science 376:156–162
Zhong S, Feng J (2022) CircPrimer 2.0: a software for annotating circRNAs and predicting translation potential of circRNAs. BMC Bioinformatics 23:215
Acknowledgements
This work was supported by the National Natural Science Foundation of China (32370151, 41376135).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Communicated by Johannes Wittmann
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Fangxin Lu and Shuang Wu contributed equally to this work.
Electronic Supplementary Material
Below is the link to the electronic supplementary material

705_2023_5908_MOESM4_ESM.jpg
Supplementary Fig. S1 Maximum-likelihood tree constructed using the RdRP palm domain region of the 126 viral sequences. The inner ring shows the taxonomic rank, and the outer ring shows the sources of the viral genomic sequences. Forty-five taxonomically undetermined viruses are indicated by stars. Bootstrap support values were calculated by 1000 iterations of tree reorganization implemented in FastTree.

705_2023_5908_MOESM5_ESM.jpg
Supplementary Fig. S2 Heat map of the pairwise proteomic similarities of the 92 viruses clustered in the proposed locarna-like clade.

705_2023_5908_MOESM8_ESM.xls
Supplementary Table S2 The estimated genome completeness of the 126 viruses analyzed and their taxonomic information in NCBI
705_2023_5908_MOESM10_ESM.xls
Supplementary Table S4 Sequence similarities of the YS-LWW RdRPs detected in different samples by using the nested RT-PCR.
705_2023_5908_MOESM11_ESM.xls
Supplementary Table S5 Sources of the 126 viral sequences analyzed in this study. The 92 viruses clustered in the proposed locarna-like clade, and the 45 suggested novel members of the genus Locarnavirus are shown in the separated sheets.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lu, F., Wu, S., Ni, Y. et al. Metavirome-assembled genome sequence of a new aquatic RNA virus expands the genus Locarnavirus. Arch Virol 168, 279 (2023). https://doi.org/10.1007/s00705-023-05908-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00705-023-05908-1