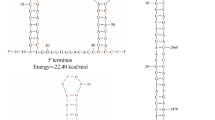
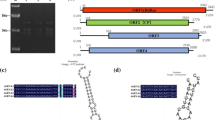
Abstract
A novel mitovirus was detected in taro (Colocasia esculenta) growing in Ningbo, China. The complete genome sequence of Colocasia esculenta associated mitovirus 1 (CeaMV1) was determined by next-generation sequencing combined with RT-PCR and RACE. The genome is 2921 nucleotides long and contains a single ORF encoding a putative RNA-dependent RNA polymerase. Homology searches and phylogenetic analysis suggested that CeaMV1 is a member of a new species in the genus Duamitovirus. This is the first report of a member of the family Mitoviridae associated with taro.


Similar content being viewed by others
Data availability
The complete genome sequence of CeaMV1 has been deposited in the NCBI database with accession number OR134096.
References
Yusop MSM, Saad MFM, Talip N, Baharum SN, Bunawan H (2019) A Review on viruses infecting taro (Colocasia esculenta (L) Schott). Pathogens (Basel, Switzerland) 8(2):56. https://doi.org/10.3390/pathogens8020056
Zettler FW (1987) Dasheen mosaic virus infecting taro in People’s Republic of China. Plant Dis 71:837–839. https://doi.org/10.1094/pd-71-0837
Gao LL, Jiang SS, Bin WU, Zhang M, Wang SJ, Xin XQ (2017) Field investigation and identification of dasheen mosaic virus on taro in Shandong province. China Veget 7:68–72. https://doi.org/10.19928/j.cnki.1000-6346.2017.07.014
Kazmi SA, Yang Z, Hong N, Wang G, Wang Y (2015) Characterization by small RNA sequencing of taro bacilliform CH virus (TaBCHV), a novel badnavirus. PLoS ONE 10(7):e0134147. https://doi.org/10.1371/journal.pone.0134147
Botella L, Manny AR, Nibert ML, Vainio E (2021) Create 100 new species and four new genera (Cryppavirales: Mitoviridae). https://ictv.global/ictv/proposals/2021.003F.R.Mitoviridae_100nsp_4ngen.zip
Walker PJ, Siddell SG, Lefkowitz EJ, Mushegian AR, Adriaenssens EM, Alfenas-Zerbini P, Dempsey DM, Dutilh BE, García ML, Curtis Hendrickson R, Junglen S, Krupovic M, Kuhn JH, Lambert AJ, Łobocka M, Oksanen HM, Orton RJ, Robertson DL, Rubino L, Sabanadzovic S, Simmonds P, Smith DB, Suzuki N, Van Doorslaer K, Vandamme AM, Varsani A, Zerbini FM (2022) Recent changes to virus taxonomy ratified by the International Committee on Taxonomy of Viruses. Arch Virol 167(11):2429–2440. https://doi.org/10.1007/s00705-022-05516-5
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30(4):772–780. https://doi.org/10.1093/molbev/mst010
Gouet P, Courcelle E (2002) ENDscript: a workflow with web interface to display sequence and structure information. Bioinformatics 18(5):767–768. https://doi.org/10.1093/bioinformatics/18.5.767
Acknowledgements
We thank Prof. M. J. Adams, Minehead, UK, for correcting the English of the manuscript.
Funding
This work was financially supported by Ningbo Major Special Projects of the Plan “Science and Technology Innovation 2025” (2021Z106), China Agriculture Research System of MOF and MARA (CARS-24-C-04), Ningbo Public Welfare Plan Project “Science and Technology Commissioner Project” (2022S203) and sponsored by the K. C. Wong Magna Fund of Ningbo University.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Handling Editor: Ioly Kotta-Loizou.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wu, M., Huang, C., Wang, L. et al. Complete genome sequence of a novel mitovirus detected in Colocasia esculenta. Arch Virol 168, 281 (2023). https://doi.org/10.1007/s00705-023-05902-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00705-023-05902-7