Abstract
Rabies virus (RABV) infection leads to a fatal neurological outcome in humans and animals and is associated with major alterations in cellular gene expression. In this study, we describe the effects of RABV infection on the mRNA expression levels of two genes, encoding the Ca2+-binding proteins (Ca-BPs) calbindin D-28K (Calb1) and calretinin (Calb2), in the brains of BALB/c mice. Sixty 4-week-old mice were divided into two test groups and one control group. Mice were inoculated intramuscularly with either a street rabies virus (SRV) strain or a challenge virus standard (CVS-11) strain and sacrificed at 3-day intervals up to day 18 postinfection. A direct fluorescent antibody test (DFAT) was used to verify the presence of RABV antigen in brain tissues, and real-time quantitative PCR (RT-PCR) was used to assess gene expression. Infection with both RABV strains resulted in significant (p < 0.05) increases in Calb1 and Calb2 expression in the test animals when compared with the controls at various time points in the study. Correlation analysis indicated very weak insignificant (p > 0.05) negative and positive relationships, respectively, between Calb1 expression (r = -0.04) and Calb2 expression (r = 0.08) with viral load (CVS-11 strain). Insignificant (p > 0.05) relationships were also observed Calb1 expression (r = -0.28) and Calb2 expression (r = 0.06) and viral load for the SRV strain.
The observed alterations in Calb1 and Calb2 expression in this study indicate possible impairments in neuronal Ca2+ buffering and Ca2+ homeostasis as a result of RABV infection and, consequently, possible involvement of calbindin-D28K and calretinin in the neuropathogenesis of rabies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rabies remains a worrying neglected tropical disease of public health significance in many developing countries, and it causes up to 60,000 deaths worldwide, annually [19]. The disease is an often-fatal zoonosis caused by rabies virus (RABV), a single-stranded, negative-sense RNA virus belonging to the genus Lyssavirus of the family Rhabdoviridae [14]. Capable of infecting almost all mammals and highly neurotropic, RABV is transmitted mainly through the bite of an infected animal and the inoculation of virus-laden saliva into the bite wound.
RABV gains access to the peripheral nervous system (PNS) from the bite site, subsequently spreading through viral replication and retrograde axonal transport until it reaches the central nervous system (CNS) and rapidly advances to the brain, where it causes an intense infection characterized by marked neurological features and an almost 100% fatality rate in untreated human patients [12, 16]. Despite the extensive investigations that have been carried out in the past, the precise basis of the functional aberrations observed in RABV-infected neurons remains to be elucidated, and there are still gaps in our understanding of the mechanisms underlying the neuropathogenesis of rabies [18, 31].
RABV infection has been shown to lead to altered levels of gene expression [27], and therefore investigating the complex mechanisms of neuronal cell dysfunction from the perspective of virus-induced changes in the expression of genes involved in the regulation of key neuronal processes such as ion homeostasis (such as genes encoding proteins involved in Ca2+ homeostasis) could be useful in expanding the currently existing knowledge about RABV neuropathogenesis. Understanding about the mechanisms of RABV pathogenesis will help to identify new molecular targets for the development of vaccines or safe therapeutics.
EF-hand proteins are a family of calcium (Ca2+)-binding proteins (Ca-BPs), which are central to cellular Ca2+ signaling by broadly functioning as Ca2+ buffers or Ca2+ sensors. These proteins share a common motif known as the EF-hand, which is involved in binding Ca2+ ions selectively and with high affinity [32, 34]. EF-hand proteins are involved in a wide array of cellular activities such as the control of Ca2+ gating, the modulation of Ca2+ signaling kinetics, and transduction of the signals into a biochemical response [11, 17].
Calbindin-D28K (CB) is an intracellular protein belonging to the EF-hand family of calcium-binding proteins that functions in part as a cytosolic calcium buffer [11, 39]. Encoded by the Calb1 gene, CB is mainly expressed by double bouquet cells (axodendritic inhibitory interneurons) as well as by some Cajal-Retzius neurons in the CB-positive neurons of the neuronal cortex in the human brain [4, 34]. In addition to its role as a cytosolic Ca2+ buffer, CB has also been postulated to function in Ca2+ transport and play a role in protection against Ca2+ overload [45]. Pathological conditions such as Alzheimer’s disease have been reported to be associated with altered patterns of CB distribution in various regions of the brain [10], suggesting that it would be worthwhile to investigate its expression in rabies.
Calretinin (CR) is another EF-hand protein that is predominantly expressed in specific neurons of the central and peripheral nervous system. Its expression may also be observed in non-neuronal cells such as mesothelial cells and during embryonic development [33]. Encoded by the Calb2 gene, CR functions as a fast Ca2+ buffer protein, altering the shape of intracellular Ca2+ transients [5]. Studies have also shown that CR undergoes considerable conformational changes upon Ca2+ binding, suggesting that it also functions as a Ca2+ sensor in addition to its Ca2+ buffering role. Although relatively little is known about the mechanisms of regulation of CR expression in various tissues, alterations in the expression levels of CR have been reported to be associated with some human diseases and also to occur as a result of experimental manipulations in animal models [33, 34].
Both CR and CB are strongly expressed in a subset of inhibitory local circuit neurons with specific functions in GABAergic neurotransmission [3]. Reports have indicated a possible involvement of this GABAergic system during RABV infection [42], and a sustained change in the expression patterns of CB and CR in the brains of RABV-infected mice could lead to alterations in Ca2+ homeostasis and impairments in GABAergic neurotransmission, potentially contributing to neuronal dysfunction in rabies [39]. Additionally, RABV can offset the balance of the Ca2+/GABA mechanism by redirecting cellular resources toward viral gene expression by promoting immune evasion [29].
In addition to the expression levels of Ca-BPs being altered in brain disease states [22], perturbations in neuronal Ca2+ regulation have been linked with the pathogenesis of various neurodegenerative disorders [34, 44]. Presently, there is very little experimental data available on the gene expression profiles of major Ca-BPs in RABV infection. In this study, we describe changes in Calb1 and Calb2 mRNA expression associated with RABV infection in mouse brain.
Materials and methods
Animals and virus strains
Sixty BALB/c mice, matched in age (3-4 weeks) and weight, were used in the study. The mice were obtained from the animal facility of the Faculty of Pharmaceutical Sciences, Ahmadu Bello University (ABU), Zaria. The mice were maintained in a high-security area under appropriate environmental and nutritional conditions, following the standards of the Animal Care Committee of Ahmadu Bello University. Experiments involving mice were performed at the animal facility of the Department of Veterinary Public Health and Preventive Medicine, ABU, Zaria. Six animals each were placed in clean, well-ventilated cages under natural lighting conditions (12 hours of sunlight and 12 hours of darkness) and fed ad libitum with broiler mash and water. The Nigerian street rabies virus (SRV470) and the challenge virus standard (CVS-11) RABV strains used were obtained from the National Veterinary Research Institute (NVRI), Jos. All personnel involved in the study received full pre-exposure vaccination before the commencement of the study.
Experimental groups of animals and virus inoculation
The viral material used for inoculation was obtained from the macerated brain of an infected suckling mouse. A 10% w/v dilution of this material was prepared in normal saline containing 200 µg of penicillin and 4 mg of streptomycin per ml [40]. For the study, the mice were divided into three groups, all were kept under similar conditions, and all were inoculated intramuscularly in the left hind limb with 0.03 ml (containing approximately 1.0 × 106 infectious particles of RABV) of a 10% w/v dilution of inoculum as described previously [30]. The experimental groups were as follows:
-
group 1: SRV-strain-infected mice
-
group 2: CVS-11-strain-infected mice
-
group 3: uninfected/control group (inoculated with a similar volume of normal saline)
Successful infection with RABV was evaluated by monitoring the mice for neurological signs and death. On days 3, 6, 9, 12, 15, and 18 postinfection (p.i.), three mice were randomly selected from each of the RABV-infected groups and sacrificed. Uninfected control mice were also randomly selected and sacrificed at the same time as the infected animals. Whole brain tissue was collected from each animal and preserved in RNA Later at - 20 °C until required.
Direct fluorescent antibody test (DFAT)
A direct fluorescent antibody test (DFAT) was carried out on the collected brain tissue samples as described by WHO [30] to verify the presence of the rabies virus antigen. Briefly, an impression smear on a clean glass slide was prepared from each brain tissue sample, air dried at room temperature, and fixed in a Coplin jar containing cold acetone at -20 °C for 30 minutes. The slides were removed from the acetone, air-dried at room temperature, and stained in a humid chamber with a fluorescein-labeled monoclonal anti-RABV immunoglobulin (Fujirebio Diagnostics, Inc., Malvern, Pennsylvania, USA). Positive (containing a previously confirmed positive tissue sample) and negative (containing a previously confirmed negative tissue sample) control slides were also prepared similarly to the experimental samples. All of the slides were then incubated at 37 °C for 30 minutes, removed from the incubator, and washed three times with 1X phosphate-buffered saline (PBS) (pH 7.4). The slides were then air-dried and visualized under a fluorescent microscope. The presence of brilliant apple green or greenish-yellow fluorescing intracellular particles or accumulations against a dark background was regarded as a positive result (Plates 1, 2).
RNA isolation and complementary DNA (cDNA) synthesis
Total RNA was extracted from 30 mg of RABV-antigen-positive mouse brain tissue using a Norgen Total RNA Purification Kit (Norgen Biotek Corp., Ontario, Canada), following the manufacturer’s protocols. RNA quality was assessed using a NanoDrop ND-1000 spectrophotometer. Following the manufacturer’s instructions, a SensiFASTTM cDNA synthesis kit (Bioline Reagents Ltd., UK) was used to transcribe total RNA to cDNA, Briefly, a 20-µl master mix containing up to 15 µl of RNA template (500 ng), 1 µl of reverse transcriptase, 4 µl of 5x TransAmp buffer containing a mixture of random hexamers and oligo dT, and variable amounts of DNase/RNase-free water per tube was prepared. PCR cycling conditions were set at 25 °C for 10 minutes, 42 °C for 15 minutes, and 85 °C for 15 minutes for a total of 35 cycles. After synthesis, the cDNA products were stored at -20 °C for further analysis.
Quantitative real-time PCR
Quantitative real-time polymerase chain reaction was performed to measure gene expression using a BIOER Line-Gene system (Hangzhou, China). Mouse phosphoglycerate kinase 1 (PGK) was used as an endogenous control (reference gene). The primers for Calb1, Calb2, and PGK are listed in Table 1. For the reaction, a master mix containing 2 µl of cDNA template (500 ng), 5 µl of 5x HOT FIREPol® Eva Green (No ROX) qPCR Mix Plus (Solis Bio-Dyne, Tartu, Estonia), 0.6 µl each of forward and reverse primers (10 µM final concentration), and 16.8 µl of nuclease-free PCR-grade water to a final volume of 25 µl per tube was prepared. Real-time PCR cycling conditions were set as follows: 95 °C for 15 minutes (initial inactivation), 40 cycles of 95 °C for 15 seconds, 63 °C for 20 seconds, and 72 °C for 20 seconds. CT (cycle threshold) values obtained from the real-time PCR data were analyzed using the 2-∆∆CT relative quantification method.
Determination of viral load
To determine the viral load of RABV in the infected brain tissue, nested PCR was carried out using 5x FIREPol® Master Mix (Solis Bio-Dyne, Tartu, Estonia), which is a ready-to-load PCR synthesis kit, following the manufacturer’s protocol. Using virus standards for the SRV and CVS-11 strains of RABV, cDNA was generated from the N gene region of each strain, using specific N gene primers (Table 2). The cDNA amplicons were purified, quantified using a NanoDrop spectrophotometer, and used to prepare tenfold serial dilutions of known concentration. Next, a set of nested primers specific for regions within the N-gene templates was then used to carry out qPCR on each of the serial dilutions to obtain Ct values. A standard curve of Ct vs. Log10 quantity was plotted from the dilutions. To generate Ct values from the test samples, qPCR was carried out on each sample using the nested PCR primers only, as shown in Table 2. Using the standard curve, the quantity of viral material in each test sample was extrapolated from its Ct value.
Data analysis
Each experiment was repeated three times to address experimental variability, and numerical results were presented as the mean ± standard deviation (mean ± SD). Data analysis was performed using the software Statistical Package for Social Sciences (SPSS) version 20.0 (SPSS Inc., Chicago, Illinois, USA). p-values less than 0.05 were considered statistically significant. Quantitative variable distribution was determined using the Shapiro-Wilk test. Variance homogeneity was determined by Levene’s test. Mean differences in expression between the test and control groups at each time point were determined using one-way analysis of variance (ANOVA). Tukey’s post hoc test was used to assess individual differences between groups. Pearson’s correlation analysis was carried out to determine the relationship between RABV titer and the Calb1 and Calb2 gene expression patterns. The sensitivity, specificity, and positive and negative predictive values of the DFAT screening results and viral load assays were determined at the 95% confidence level using SPSS. Using the serial dilutions with which the standard curve was plotted, the limits of detection of the qPCR assay for measuring the viral load were determined for the SRV and CVS-11 strains of RABV using a Microsoft Excel data analysis function.
Results
In this study, screening by DFAT (Plates 1, 2) confirmed the presence of RABV antigen in 83.3% (20/24) and 88% (21/24) of the mouse brain samples inoculated with street rabies virus (SRV) and challenge virus standard 11 (CVS-11) strains, respectively (Table 3). Clinical signs of rabies in animals such as ruffled fur, paralysis in one or more of the limbs, and loss of mobility were observed in the infected animals from the fifth day p.i.
To assess Calb1 mRNA expression levels in the brain tissues of SRV-infected mice, CVS-11-infected mice, and uninfected control mice, quantitative real-time PCR was performed. One-way ANOVA was used to analyze the differences in expression between the various groups at each measured time point.
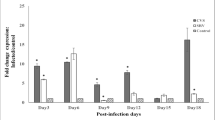
Results from the gene expression studies (Fig. 1) showed that infection with either the SRV or CVS-11 strain of RABV resulted in similar increases in Calb1 mRNA levels throughout the study period except for day 6, on which a notable difference in the effect of the two strains was observed. There was a significant difference between the SRV, CVS-11, and control groups on day 3 p.i., as determined by one-way ANOVA (F (2, 6) = 7.265, p = 0.025). A Tukey HSD post hoc test for multiple comparisons revealed that Calb1 expression was significantly higher in the CVS-11 group (12.30 ± 4.10-fold) than that in the control group (p = 0.026). There was no statistically significant difference between the expression in the SRV group and the CVS-11 group (p = 0.067) or the SRV group and the control group (p = 0.721) for the same day.
Expression of Calb1 mRNA (fold change normalized to PGK-1) in the brain tissue of RABV-infected mice. Mean values with different letters show a statistically significant difference when compared with others on the same day postinfection (p < 0.05) .SRV, street rabies virus–infected mice; CVS, challenge virus standard-11–infected mice; Calb1, calbindin-D28K gene; CTRL, uninfected control mice; PGK-1 phosphoglycerate kinase 1
On day 6 p.i., ANOVA indicated a significant difference in Calb1 expression between the SRV, CVS-11, and control groups (F (2, 6) = 6.865, p = 0.028). Post hoc analysis using Tukey’s HSD test for multiple comparisons showed that there was a significant difference (p = 0.033) between the Calb1 levels in SRV-infected mice (5.80 ± 2.22-fold expression) and those in CVS-11-infected mice (0.87 ± 0.44-fold) on the same day.
On day 9 p.i., infection with both RABV strains had significant effects on Calb1 expression (F (2, 6) = 6.116, p = 0.036). Tukey’s post hoc test revealed statistically significantly increased Calb1 levels in the CVS-11 group (3.22 ± 2.18-fold expression) compared to the control group (p = 0.031). On days 12, 15, and 18 p.i., ANOVA did not reveal any significant differences in Calb1 expression between the groups.
To assess Calb2 mRNA expression levels in the brain tissues of SRV-infected mice, CVS-11-infected mice, and uninfected control mice, quantitative real-time PCR was performed. A one-way ANOVA was performed to compare the effects of RABV infection on the three groups. The results showed that infection with RABV led to alterations in Calb2 expression levels, which were significant on days 6 and 9 p.i (Fig. 2).
Calb2 expression levels differed significantly between the groups on day 6 p.i. (F (2, 6) = 23.975, p = 0.001). Multiple comparisons using Tukey’s HSD post hoc test indicated significantly increased (p = 0.015) Calb2 expression (10.87 ± 7.27-fold) in the SRV group compared to the control group for the same day. Unlike the increased expression in the SRV group, CVS-11-infected mice showed a decrease in Calb2 levels (0.42 ± 0.04-fold expression), which were significantly different from those of the SRV group for the day (p = 0.001). There was no significant difference in expression between the CVS-11 and control group on day 6 p.i. (p = 0.069).
ANOVA indicated a significant difference between the groups on day 9 p.i. (F (2, 6) = 10.021, p = 0.012). Post hoc multiple comparisons of the data between the groups revealed a significant increase in Calb2 levels (2.14 ± 0.80-fold expression) in the SRV group compared to the control group on the same day (p = 0.012). There was also a significant difference between the Calb2 levels in the SRV group and the CVS-11 group (2.05 ± 1.08-fold expression) on day 9 p.i. (p = 0.43). There was no significant difference between the Calb2 expression levels in the CVS-11 and control groups on day 9 p.i. (p = 0.542).
Data analysis revealed no statistically significant differences between the expression in the SRV, CVS-11, and control groups at other measured time points in the study (days 3, 12, 15, and 18 p.i).
Relationship between mRNA expression and viral loads in RABV-infected mice
To investigate possible relationships between rabies viral loads and gene expression level changes in the brain tissues of SRV- and CVS-11-infected mice, nested PCR was performed to determine the viral load (Table 4), and Pearson’s correlation analysis was then performed to correlate the variables. In SRV-infected mice (Table 5), a weak negative correlation (r = -0.288) was observed between Calb1 expression and viral load. For Calb2 expression, a weak positive correlation (r = 0.066) with viral load was observed. There was no significant association (p > 0.05) between the changes in expression of either Calb1 or Calb2 and viral load (Table 5). In CVS-11-infected mice (Table 5), Calb1 expression showed a weak negative correlation (r = -0.044) with viral load, while Calb2 expression showed a weak positive correlation (r = 0.088) with viral load. The association between the changes in expression of both Calb1 and Calb2 and viral load in CVS-11-infected mice was also not significant (p > 0.05).
In SRV-infected mice, sensitivity and specificity estimates of 87.0% (95% confidence interval [CI], 0.70-0.96) and 85.7% (95% CI, 0.67-0.95), respectively, were obtained. At a 95% CI, the positive predictive value (PPV) and negative predictive value (NPV) estimates for SRV-infected mice were 90.0% (95% CI, 0.75-0.98) and 80% (95% CI, 0.56-0.95), respectively. The detection limit of the qPCR assay to determine the viral load in SRV-infected mice was found to be 20 cDNA copies/µl.
For CVS-11-infected mice, sensitivity and specificity estimates of 87.5% (95% CI, 0.71-0.96) and 85.7% (95% CI, 0.63-0.97), respectively, were obtained. The positive predictive value (PPV) and negative predictive value (NPV) estimates for CVS-11-infected mice were 91% (95% CI, 0.76-0.98) and 80% (95% CI, 0.57-0.95), respectively. The detection limit of the qPCR assay to determine the viral load in CVS-11-infected mice was found to be 22 cDNA copies/µl.
Discussion
In an attempt to understand the possible roles played by the Ca2+ buffer proteins calbindin and calretinin in rabies neuropathogenesis, the expression levels of their genes were examined over time. Our study reports for the first time the comparative effects of rabies virus infection on the mRNA expression patterns of the Calb1 and Calb2 genes in the brain tissues of mice infected with the SRV and CVS-11 strains of RABV. The alterations in Calb1 and Calb2 expression reported in this study corroborate earlier reports that RABV infection elicits a cellular response, leading to differential expression of various genes and proteins in the CNS, especially neurotransmitter-associated genes [27, 41]. Processes related to neuronal function have been reported to be altered very early in infection, before an immune response is induced [18], in agreement with the results of this study.
For both genes that were investigated in the study, higher levels of expression were observed at different time points than in the control group. Although the changes in Calb1 and Calb2 expression levels generally differed only slightly between the two virus strains in this study, a notable contrast between the strains was observed on day 6 p.i. While SRV-infected mice showed an increased expression of 5.8-fold for Calb1, CVS-11-infected mice showed a decreased expression of 0.9-fold for the same gene. For Calb2, SRV-infected mice showed a 6-fold increase in expression, and a 0.4-fold decrease in expression was observed in CVS-11-infected mouse brain.
We postulate that the increase in Calb1 expression (especially on days 3-6 p.i.) was due to a response by the host to limit neuronal dysfunction or death. Previous studies have shown that activation of the interferon system leads to the establishment of antiviral responses that help to shape an effective response [1, 25]. A cellular response mechanism to cope with a possible sudden imbalance in cytosolic Ca2+ levels caused by SRV and CVS-11 infection could have led to the increase in gene expression observed in the study. Mitochondria and the endoplasmic reticulum (ER) can accumulate cytosolic Ca2+ [46], and it is possible that a disruption in Ca2+ release or storage from these intracellular stores could result from RABV infection as a result of organelle dysfunction. Jackson et al. and Alandijany et al. have reported mitochondrial dysfunction as a result of RABV infection [47, 48], which agrees with our postulation. RABV has been demonstrated to utilize endosomal systems of host cells for internalization and trafficking [35]. In addition, the downregulation of endoplasmic proteins that regulate membrane transport has been shown to significantly reduce RABV infection, suggesting that these proteins may play important roles during RABV infection [2].
The increased expression in this study could also have been due to viral replication processes interfering with the normal regulation of Calb1 gene expression. Calbindin is a cytoplasmic protein, and RABV replication occurs in the cytoplasm of the cell [23]. Interference with the translation of Calb1 mRNA due to virus replication in the cell cytoplasm could possibly initiate mechanisms leading to increased gene transcription. The eventual suppression of Calb1 expression in the later stages of the study (days 9-18 p.i.) as compared to the earlier stages (days 3-6 p.i.) could also have been due to viral replication interference. Hence, we attribute the differences in expression between the different stages of the study to the possible interference of virus replication with normal gene regulation. This is supported by experimental reports that RABV infection does not induce immediate cell death or degradation in neurons but leads to neuronal dysfunction via the alteration of host gene expression [9]. Additionally, the onset of clinical signs (increased agitation and gradual paralysis) in the test animals corresponds to the period of reduced gene expression in the study.
The increased Calb2 expression observed in our study might be attributable to the same causes as the increased expression of Calb1, especially since these two genes encode proteins with closely related functions [11, 13]. Since calretinin expression is normally regulated at the post-transcriptional level by the transcription factors NRF-1 and E2F2 [20, 21], the probability that cytoplasmic RABV replication interferes with the normal regulation of Calb2 expression is further strengthened.
Although some studies have reported a loss of calbindin expression in the brains of animal models with rabies [39, 42], upregulation of gene expression as an outcome of RABV infection in animal models has also been reported. Increased parvalbumin expression has been observed in the cerebral cortex of mice infected with RABV [40]. Intracerebral infection of mice with the CVS-N2c strain of RABV was shown by Prosniak et al. to result in upregulation in the expression of 1.4% of the genes evaluated in that study [27]. Some genes were downregulated early in the study and then upregulated in the later stages (days 6 and 7 p.i.), while a few were upregulated throughout the entire study period. Upregulated genes included genes involved in antiviral growth, cell defense, immune regulation, and the establishment of new connections between neurons.
Infection of suckling mice with street rabies virus has been shown to result in an increase in the expression of eight major groups of genes in the brain, as determined on different days postinfection using cDNA arrays and qPCR [41]. Upregulated gene groups included genes involved in immune response, metabolism, kinases, proteases, enzymes, death-related proteins, transcription, and translation. Experimental infection with two types (fixed and street) of RABV caused increased immunoreactivity of microtubule-associated protein 2 (MAP-2) and neurofilament (NF-H) in the spinal cord and cerebral cortex of mice [15, 24].
Koraka et al. reported the upregulation of 409 genes and downregulation of 22 genes in mouse brains by RABV-P infection, compared to 262 upregulated and eight downregulated genes by Duvenhage virus. Although RABV infection has been demonstrated to lead to an increase in the expression of some genes in mouse brain, no genes coding for calcium-binding proteins were evaluated in any of the studies in which upregulation was observed [19].
Although the findings from this study appear to be consistent with some of the general reports on RABV infection in animal models, there are some discrepancies between our results and those reported by Torres-Fernandez et al. and Verdes et al. [39, 42]. One reason for this might be that the genes of interest in this study were assessed at the level of transcription only; consequently, the possibility of a discrepancy between transcription and translation levels may come into play. Another explanation is that whole-brain lysates (total brain mRNA) were used for this study, whereas samples from specific brain regions (such as the cerebral cortex and the hippocampus) were used in other studies. Natural infection with rabies virus has been observed to cause a loss in CB immunostaining of Purkinje cells in the cerebellar cortex of cattle, while the large interneurons in the granular layer were observed to maintain their positive immunostaining [42]. From this, it is reasonable to infer that while Ca-BP expression can be downregulated in some brain regions, it can remain the same or even increase in other regions.
The route of inoculation could have also played a role, as different routes of inoculation have been associated with different effects in the brains of mice inoculated with RABV. Previous studies have shown that intracerebral inoculation of mice with RABV caused apoptosis, while intramuscular inoculation did not [9, 28]. Since intracerebral inoculation eliminates the need for the virus to travel to the brain, it may not be an appropriate way to model the processes that occur during infection, whereas intramuscular inoculation might more closely simulate what happens during natural infection. Intramuscular infection with RABV might simulate a natural infection and thus offer a more realistic approach to understanding the physio-pathological mechanisms involved [7].
The weak correlations between viral load and gene expression in this study agree with the findings from other studies that gene expression does not always reflect variations in viral load in some viral diseases [6, 43]. Overall, changes in global RNA expression mostly reflect responses to viral replication rather than a mechanism that might explain viral control. Therefore, the correlation of a gene’s expression with viral load may suggest that the changes in expression are a response to the amount of virus present or that the gene directly controls the viral load.
Impairment in Ca2+ homeostasis seems to be a common underlying factor in the pathogenic mechanism of neurodegenerative disorders such as Alzheimer’s disease, Parkinson’s disease, and amyotrophic lateral sclerosis, despite intrinsic differences in their etiologies [8, 26, 36]. This implies that sustained alterations in Ca2+ homeostasis play a major role in either the initiation or progression of neuro-dysfunctional processes by increasing the vulnerability of neuronal cells to metabolic and other stressors [37, 38]. Although the functional relevance of the upregulation of Calb1 and Calb2 expression to rabies pathogenesis requires further study, our study shows that expression of Calb1 and Calb2 is altered by infection with the SRV and CVS-11 strains of RABV.
In conclusion, expression of both Calb1 and Calb2 is altered by RABV infection. These alterations imply the involvement of Ca-BPs during RABV infection, reflecting possible impairments in the Ca2+ buffering system and regulation of Ca2+ homeostasis, with associated neuronal cell dysfunction. The varying changes in mRNA expression levels of the genes throughout the infection period indicate that the proteins they encode may be important in the correction of neuronal Ca2+ dyshomeostasis caused by RABV infection. Therefore, Calb1 and Calb2 may play significant roles at the early stages of the neuronal cell response to RABV infection and consequently are implicated in the neuropathogenesis of the disease. Relevant signaling pathways, varying host cellular responses, and pathogeny still provide vast areas in which to conduct intensive investigations regarding rabies pathogenesis.
Data availability
The datasets generated and/or analyzed during the current study are available from the corresponding author upon reasonable request.
References
Abdulazeez M, Kia GSN, Abarshi MM, Muhammad A, Ojedapo CE, Atawodi JC, Dantong D, Kwaga JKP (2020) Induction of rabies virus infection in mice brain may up and down-regulate type II interferon-gamma via epigenetic modifications. Metab Brain Dis 35:819–827. https://doi.org/10.1007/s11011-020-00553-y
Ahmad W, Li Y, Guo Y, Wang X, Ming D, Guan Z et al (2017) Rabies virus co-localizes with early (Rab5) and late (Rab7) endosomal proteins in neuronal and SH-SY5Y cells. Virol Sin 32:207–215. https://doi.org/10.1007/s12250-017-3968-959:665-677,2010
Barinka F, Druga R (2010) Calretinin expression in the mammalian neocortex: a review. Physiol Res 59(5):665–677
Benes FM, Berretta S (2001) GABAergic interneurons: implications for understanding schizophrenia and bipolar disorder. Neuropsychopharmacology 25(1):1–2. https://doi.org/10.1016/S0893-133X(01)00225-1
Blum W, Pecze L, Rodriguez JW, Steinauer M, Schwaller B (2018) Regulation of calretinin in malignant mesothelioma is mediated by septin 7 binding to the CALB2 promoter. BMC Cancer 18(1):475. https://doi.org/10.1186/s12885-018-4385-7
Bosinger SE, Hosiawa KA, Cameron MJ, Persad D, Ran L, Xu L et al (2004) Gene expression profiling of host response in models of acute HIV infection. J Immunol 173(11):6858–6863. https://doi.org/10.4049/jimmunol.173.11.6858
Consales CA, Bolzan VL (2007) Rabies review: immunopathology, clinical aspects, and treatment. J. Venom. Anim. Toxins Incl. Trop. Dis 13(1): 5-38
Duda J, Pötschke C, Liss B (2016) Converging roles of ion channels, calcium, metabolic stress, and activity pattern of Substantia nigra dopaminergic neurons in health and Parkinson’s disease. J Neurochem 139:156–178. https://doi.org/10.1111/jnc.13572
Fu ZF, Jackson AC (2005) Neuronal dysfunction and death in rabies virus infection. J Neuro Virol 11(1):101–106. https://doi.org/10.1080/13550280590900445
Geula C, Bu J, Nagykery N, Scinto LF, Chan J, Joseph J et al (2003) Loss of Calbindin-D28k from aging human cholinergic basal forebrain: relation to neuronal loss. J Comp Neurol 455(2):249–259. https://doi.org/10.1002/cne.10475
Girard F, Venail J, Schwaller B, Celio MR (2015) The EF-hand Ca2+-binding protein super-family: a genome-wide analysis of gene expression patterns in the adult mouse brain. Neurosci 294:116–155. https://doi.org/10.1016/j.neuroscience.2015.02.018
Gluska S, Zahavi EE, Chein M, Gradus T, Bauer A et al (2014) Rabies virus hijacks and accelerates the p75NTR retrograde axonal transport machinery. PLoS Pathog 10(8):e1004348. https://doi.org/10.1371/journal.ppat.1004348
Groves P, Palczewska M (2001) Cat-ion binding properties of Calretinin, an EF-hand calcium-binding protein. Acta Biochim Pol 48(1):113–119. https://doi.org/10.18388/abp.20015117
Guo Y, Ahmad W, Song Y, Wang X, Gao J, Duan M et al (2019) In vitro infection of the street and fixed rabies virus strains inhibit gene expression of actin-microtubule binding proteins EB3 and p140cap in Neurons. Pak Vet J 39(3):359–364. https://doi.org/10.29261/pakvetj/2019.007
Hurtado AP, Rengifo AC, Torres-Fernández O (2015) Immunohistochemical overexpression of MAP-2 in the cerebral cortex of rabies-infected mice. Int J Morphol 33(2):465–470. https://doi.org/10.4067/S0717-95022015000200010
Jackson AC (2014) Rabies. Handb Clin Neurol 123:601–618. https://doi.org/10.1016/b978-0-444-53488-0.00029-8
Kelemen K, Szilágyi T (2021) New approach for untangling the role of uncommon calcium-binding proteins in the central nervous system. Brain Sci 11(5):634. https://doi.org/10.3390/brainsci11050634ht
Kim S, Larrous F, Varet H, Legendre R, Feige L, Dumas G et al (2021) Early transcriptional changes in rabies virus-infected neurons and their impact on neuronal functions. Front Microbiol 12:730892–730892. https://doi.org/10.3389/fmicb.2021.730892
Koraka P, Martina BE, Van Den Ham HJ, Zaaraoui-Boutahar F, Van IJcken, W, et al (2018) Analysis of mouse brain transcriptome after experimental duvenhage virus infection shows activation of innate immune response and pyroptotic cell death pathway. Front Microbiol 9:397. https://doi.org/10.3389/fmicb.2018.00397
Kresoja-Rakic J, Kapaklikaya E, Ziltener G, Dalcher D, Santoro R, Christensen BC et al (2016) Identification of cis- and trans-acting elements regulating calretinin expression in mesothelioma cells. Oncotarget 7(16):21272–21286. https://doi.org/10.18632/oncotarget.7114
Kresoja-rakic J, Sulemani M, Kirschner MB, Ronner M, Reid G, Kao S et al (2017) Post-transcriptional regulation controls calretinin expression in malignant pleural mesothelioma. Front Genet 8(70):1–12. https://doi.org/10.3389/fgene.2017.00070
Kreutz MR, Naranjo JR, Koch KW, Schwaller B (2012) The neuronal functions of EF-hand Ca2+-binding proteins. Front Mol Neurosci. https://doi.org/10.3389/fnmol.2012.00092
Mazarakis ND, Azzouz M, Rohll JB, Ellard FM, Wilkes FJ, Olsen AL et al (2001) Rabies virus glycoprotein pseudo-typing of lentiviral vectors enables retrograde axonal transport and access to the nervous system after peripheral delivery. Human Mol Genet 10(19):2109–2121. https://doi.org/10.1093/hmg/10.19.2109
Monroy-Gómez J, Santamaría G, Torres-Fernández O (2018) Overexpression of MAP2 and NF-H associated with dendritic pathology in the spinal cord of mice infected with rabies virus. Viruses 10(3):112. https://doi.org/10.3390/v10030112
Ojedapo CE, Muhammad A, Kia GS, Abarshi MM, Abdulazeez M, Atawodi JC, Kwaga JK (2020) Rabies virus induction in mice upregulates B7–H1 via epigenetic modifications. Virus Dis. https://doi.org/10.1007/s13337-020-00588-w
Popugaeva E, Pchitskaya E, Bezprozvanny I (2017) Dysregulation of neuronal calcium homeostasis in Alzheimer’s disease–a therapeutic opportunity. Biochem Biophys Res Comm 483(4):998–1004. https://doi.org/10.1016/j.bbrc.2016.09.053
Prosniak M, Hooper DC, Dietzschold B, Koprowski H (2001) Effect of rabies virus infection on gene expression in mouse brain. PNAS 98(5):2758–2763. https://doi.org/10.1073/pnas.051630298
Rengifo A, Umbarila V, Janeth Garzón M, Torres-Fernández O (2016) Differential effect of the route of inoculation of rabies virus on NeuN immunoreactivity in the cerebral cortex of mice. Int J Morphol 34(4):1362–1368. https://doi.org/10.4067/S0717-95022016000400031
Rivas HG, Schmaling SK, Gaglia MM (2016) Shutoff of host gene expression in influenza a virus and herpes viruses: similar mechanisms and common themes. Viruses 8(4):102. https://doi.org/10.3390/v8040102
Rupprecht CE, Fooks AR, Abela-Ridder B (2018) Laboratory techniques in rabies, vol 1, 5th edn. World Health Organization, Geneva
Schutsky K, Portocarrero C, Hooper DC, Dietzschold B, Faber M (2014) Limited brain metabolism changes differentiate between the progression and clearance of rabies virus. PLoS ONE 9(4):e87180. https://doi.org/10.1371/journal.pone.0087180
Schwaller B (2009) The continuing disappearance of “pure” Ca2+ buffers. Cell Mol Life Sci 66(2):275–300. https://doi.org/10.1007/s00018-008-8564-6
Schwaller B (2014) Calretinin: from a “simple” Ca2+ buffer to a multifunctional protein implicated in many biological processes. Front Neuroanat. https://doi.org/10.3389/fnana.2014.00003
Schwaller B (2020) Cytosolic Ca2+ buffers are inherently Ca2+ signal modulators. Cold Spring Harbor Perspect Biol 12(1):a035543. https://doi.org/10.1101/cshperspect.a035543
Semerdjieva S, Shortt B, Maxwell E, Singh S, Fonarev P, Hansen J et al (2008) Coordinated regulation of AP2 uncoating from clathrin-coated vesicles by rab5 and hRME-6. J. J Cell Biol. 183:499–511. https://doi.org/10.1083/jcb.200806016
Sirabella R, Valsecchi V, Anzilotti S, Cuomo O, Vinciguerra A et al (2018) Ionic homeostasis maintenance in ALS: focus on new therapeutic targets. Front Neurosci 12:510. https://doi.org/10.3389/fnins.2018.00510
Toescu EC, Verkhratsky A (2004) Ca2+ and mitochondria as substrates for deficits in synaptic plasticity in normal brain aging. J Cell Mol Med 8(2):181–190. https://doi.org/10.1111/j.1582-4934.2004.tb00273.x
Toescu EC, Vreugdenhil M (2010) Calcium and normal brain aging. Cell Calcium 47(2):158–164. https://doi.org/10.1016/j.ceca.2009.11.013
Torres-Fernandez O, Yepes GE, Gomez JE, Pimenta HJ (2005) Calbindin distribution in cortical and subcortical brain structures of normal and rabies-infected mice. Int J Neurosci 115(10):1375–1382. https://doi.org/10.1080/00207450590956396
Torres-Fernández O, Yepes GE, Gómez JE, Pimienta HJ (2004) Effect of rabies virus infection on the expression of Parvalbumin, Calbindin and Calretinin in mouse cerebral cortex. Biomedica 24(1):63–78
Ubol S, Kasisith J, Mitmoonpitak C, Pitidhamabhorn D (2006) Screening of upregulated genes in suckling mouse central nervous system during the disease stage of rabies virus infection. Microbiol Immunol 50(12):951–959. https://doi.org/10.1111/j.1348-0421.2006.tb03871.x
Verdes JM, de Sant’Ana FJF, Sabalsagaray MJ, Okada K, Calliari A, Moraña JA, de Barros CSL (2016) Calbindin D28k distribution in neurons and reactive gliosis in cerebellar cortex of natural Rabies virus–infected cattle. J Vet Diag Invest 28(4):361–368. https://doi.org/10.1177/1040638716644485
Wieland S, Makowska Z, Campana B, Calabrese D, Dill MT, Chung J et al (2014) Simultaneous detection of hepatitis C virus and interferon-stimulated gene expression in infected human liver. Hepatology 59(6):2121–2130. https://doi.org/10.1002/hep.26770
Wojda U, Salinska E, Kuznicki J (2008) Calcium ions in neuronal degeneration. IUBMB Life 60(9):575–590. https://doi.org/10.1002/iub.91
Yoo YM, Jeung EB (2018) Calbindin-D28k in the brain influences the expression of the cellular prion protein. Oxi Med Cell Long. https://doi.org/10.1155/2018/4670210
Brini M, Calì T, Ottolini D, Carafoli E (2014) Neuronal calcium signaling: function and dysfunction. Cell Mol Life Sci 71(15):2787–2814
Jackson AC, Alandijany T, Kammouni W, Chowdhury SR, Fernyhough P (2013) Rabies virus infection is associated with mitochondrial dysfunction. J Neurol Sci 333:e609
Alandijany T, Kammouni W, Roy Chowdhury SK, Fernyhough P, Jackson AC (2013) Mitochondrial dysfunction in rabies virus infection of neurons. J Neurovirol 19(6):537–549. https://doi.org/10.1007/s13365-013-0214-6
Acknowledgements
The authors are grateful to the Africa Centre of Excellence for Neglected Tropical Diseases and Forensic Biotechnology, (ACENTDFB), Ahmadu Bello University (ABU) Zaria, for funding the study. The authors are also grateful to the National Veterinary Research Institute (NVRI) Jos, for providing the virus strains used in this study.
Funding
This study was funded by the African Center of Excellence for Neglected Tropical Diseases and Forensic Biotechnology (ACENTDFB), Ahmadu Bello University (ABU) Zaria, as part of graduate study funding for George C Korie.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study’s conception and design. Material preparation, data collection, and analysis were performed by George C. Korie, Brenda Kanu, and Grace S.N. Kia. Supervision was performed by Abdullahi B. Sallau and Jacob Kwaga. The first draft of the manuscript was written by George C. Korie, and all authors commented on previous versions of the manuscript. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Ethical approval
Animal housing and experimental procedures were performed according to the guidelines of the Nigerian Council on Animal Care with the approval of the Ahmadu Bello University Animal Care Committee (ABUACC).
Additional information
Handling Editor: William G Dundon.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Korie, G.C., Sallau, A.B., Kanu, B. et al. Rabies virus infection is associated with variations in calbindin D-28K and calretinin mRNA expression levels in mouse brain tissue. Arch Virol 168, 143 (2023). https://doi.org/10.1007/s00705-023-05753-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00705-023-05753-2