Abstract
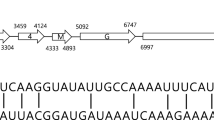
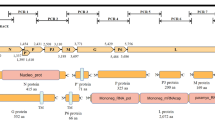
Through high-throughput RNA sequencing, we discovered a putative new cytorhabdovirus in the seeds of Rudbeckia sp., which we have tentatively named “rudbeckia virus 1” (RudV1). Its complete 12,502-nt genomic sequence contains five open reading frames (ORFs): ORF1 (putative nucleocapsid protein, N), ORF2 (putative phosphoprotein, P), ORF3 (putative cell-to-cell movement protein, P3), ORF4 (putative matrix protein, M), and ORF5 (putative RNA-dependent RNA polymerase, L). BLASTp searches showed that ORF1, ORF3, ORF4, and ORF5 of RudV1 are most closely related to the corresponding proteins of Tagetes erecta virus 1 (a putative member of the genus Cytorhabdovirus) with 33.87% (88% query coverage), 55.98% (89% query coverage), 35.33% (94% query coverage), and 57.75% (98% query coverage) sequence identity at the amino acid level, respectively. Phylogenetic analysis and pairwise comparisons indicated that RudV1 is a novel member of the genus Cytorhabdovirus within the family Rhabdoviridae.


Similar content being viewed by others
References
Kuzmin IV, Novella IS, Dietzgen RG, Padhi A, Rupprecht CE (2009) The rhabdoviruses: biodiversity, phylogenetics, and evolution. Infect Genet Evol 9(4):541–553. https://doi.org/10.1016/j.meegid.2009.02.005
Dietzgen RG, Bejerman NE, Goodin MM, Higgins CM, Huot OB, Kondo H, Martin KM, Whitfield AE (2020) Diversity and epidemiology of plant rhabdoviruses. Virus Res 281:197942. https://doi.org/10.1016/j.virusres.2020.197942
Jackson AO, Dietzgen RG, Goodin MM, Bragg JN, Deng M (2005) Biology of plant rhabdoviruses. Annu Rev Phytopathol 43:623–660. https://doi.org/10.1146/annurev.phyto.43.011205.141136
Bejerman N, Dietzgen RG, Debat H (2021) Illuminating the plant rhabdovirus landscape through metatranscriptomics data. Viruses 13(7):1304. https://doi.org/10.3390/v13071304
Christie SR, Christie RG, Edwardson JR (1974) Transmission of a bacilliform virus of sowthistle and Bidens pilosa. Phytopathology 64(6):840–845
Dietzgen RG, Callaghan B, Campbell PR (2007) Biology and genomics of lettuce necrotic yellows virus. Plant Viruses 1(1):85–92
Heim F, Lot H, Delecolle B, Bassler A, Krczal G, Wetzel T (2008) Complete nucleotide sequence of a putative new cytorhabdovirus infecting lettuce. Arch Virol 153(1):81–92. https://doi.org/10.1007/s00705-007-1071-5
Liu Q, Jin J, Yang L, Zhang S, Cao M (2021) Molecular characterization of a novel cytorhabdovirus associated with chrysanthemum yellow dwarf disease. Arch Virol 166(4):1253–1257. https://doi.org/10.1007/s00705-021-04987-2
Peters D, Kitajima EW (1970) Purification and electron microscopy of sowthistle yellow vein virus. Virology 41(1):135–150. https://doi.org/10.1016/0042-6822(70)90061-9
Redinbaugh MG, Hogenhout SA (2005) Plant rhabdoviruses. Curr Top Microbiol Immunol 292:143–163. https://doi.org/10.1007/3-540-27485-5_7
Salem N, Odeh S, Muslem MA, Tahzima R (2020) Occurrence and partial genetic characterisation of Lettuce big-vein associated virus and Mirafiori lettuce big-vein virus infecting lettuce in Jordan. Ann Appl Biol 177(1):90–100. https://doi.org/10.1111/aab.12595
Whitfield AE, Huot OB, Martin KM, Kondo H, Dietzgen RG (2018) Plant rhabdoviruses—their origins and vector interactions. Curr Opin Virol 33:198–207. https://doi.org/10.1016/j.coviro.2018.11.002
Dietzgen RG, Kondo H, Goodin MM, Kurath G, Vasilakis N (2017) The family Rhabdoviridae: mono- and bipartite negative-sense RNA viruses with diverse genome organization and common evolutionary origins. Virus Res 227:158–170. https://doi.org/10.1016/j.virusres.2016.10.010
Yan T, Zhu JR, Di D, Gao Q, Zhang Y, Zhang A, Yan C, Miao H, Wang XB (2015) Characterization of the complete genome of Barley yellow striate mosaic virus reveals a nested gene encoding a small hydrophobic protein. Virology 478:112–122. https://doi.org/10.1016/j.virol.2014.12.042
Jun M, Shin S, Lee SJ, Lim S (2021) First report of Raphanus sativus cryptic virus 2 on cabbage seeds (Brassica rapa subsp. pekinensis) imported from Italy. Virus Dis 32(4):834–836. https://doi.org/10.1007/s13337-021-00744-w
Higgins CM, Bejerman N, Li M, James AP, Dietzgen RG, Pearson MN, Revill PA, Harding RM (2016) Complete genome sequence of Colocasia bobone disease-associated virus, a putative Cytorhabdovirus infecting taro. Arch Virol 161(3):745–748. https://doi.org/10.1007/s00705-015-2713-7
Tanno F, Nakatsu A, Toriyama S, Kojima M (2000) Complete nucleotide sequence of Northern cereal mosaic virus and its genome organization. Arch Virol 145(7):1373–1384. https://doi.org/10.1007/s007050070096
Tamura K, Stecher G, Kumar S (2021) MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol 38(7):3022–3027. https://doi.org/10.1093/molbev/msab120
Zhang S, Huang A, Zhou X, Li Z, Dietzgen RG, Zhou C, Cao M (2021) Natural defect of a plant rhabdovirus glycoprotein gene: a case study of virus-plant coevolution. Phytopathology 111(1):227–236. https://doi.org/10.1094/PHYTO-05-20-0191-FI
Acknowledgements
This work was supported by a research fund (project no. B-1543086-2021-22-01) from the Animal and Plant Quarantine Agency, Ministry of Agriculture, Food and Rural Affairs, Republic of Korea. We would like to thank Editage (www.editage.co.kr) for English language editing.
Author information
Authors and Affiliations
Contributions
Seungmo Lim and Seong-Jin Lee contributed to the study conception and design. Experiments, data collection, and analysis were performed by Da-Som Lee, Juhyun Kim, Minji Jun, Sanghyun Shin, and Seungmo Lim. Da-Som Lee and Seungmo Lim wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Ethical approval
This article does not describe any studies involving human participants or animals performed by any of the authors.
Additional information
Handling Editor: Ralf Georg Dietzgen.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lee, DS., Kim, J., Jun, M. et al. Complete genome sequence of a putative novel cytorhabdovirus isolated from Rudbeckia sp.. Arch Virol 167, 2381–2385 (2022). https://doi.org/10.1007/s00705-022-05556-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-022-05556-x