Abstract
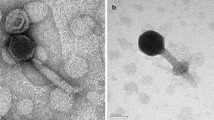
The freshwater cyanophage Mwe-Yong1112-1 was isolated using Microcystis wesenbergii as a host and found to have an icosahedral head, about 45 nm in diameter, and a flexible tail, approximately 133 nm in length and 4.5 nm in width. The complete genome of the cyanophage is 39,679 bp in length with a G+C content of 66.6%. Mwe-Yong1112-1 shared the highest pairwise average nucleotide identity (ANI) value of 67.7% (below the ≥95% boundary to define a species) and the highest nucleotide sequence similarity of 17.48% (below the >70% boundary to define a genus) with the most closely related phage. In a proteomic tree, Mwe-Yong1112-1 and three unclassified phages formed a monophyletic clade between the families Saparoviridae and Pyrstoviridae, but Mwe-Yong1112-1 occupied a separate branch from the other three phages, suggesting that it represents a new evolutionary lineage. This study enriches the available information about freshwater cyanophages.


Similar content being viewed by others
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
References
Huisman J, Codd GA, Paerl HW, Ibelings BW, Verspagen J, Visser PM (2018) Cyanobacterial blooms. Nat Rev Microbiol 16(8):471–483. https://doi.org/10.1038/s41579-018-0040-1
Harke MJ, Steffen MM, Gobler CJ, Otten TG, Wilhelm SW, Wood SA, Paerl HW (2016) A review of the global ecology, genomics, and biogeography of the toxic cyanobacterium, Microcystis spp. Harmful Algae 54:4–20. https://doi.org/10.1016/j.hal.2015.12.007
Huo D, Gan N, Geng R, Cao Q, Song L, Yu G, Li R (2021) Cyanobacterial blooms in China: diversity, distribution, and cyanotoxins. Harmful Algae 109:102106. https://doi.org/10.1016/j.hal.2021.102106
Sullivan MB, Waterbury JB, Chisholm SW (2003) Cyanophages infecting the oceanic cyanobacterium Prochlorococcus. Nature 424(6952):1047–1051. https://doi.org/10.1038/nature01929
Xia H, Li T, Deng F, Hu Z (2013) Freshwater cyanophages. Virol Sin 28(5):253–259. https://doi.org/10.1007/s12250-013-3370-1
Ou T, Li S, Liao X, Zhang Q (2013) Cultivation and characterization of the MaMV-DC cyanophage that infects bloom-forming cyanobacterium Microcystis aeruginosa. Virol Sin 28(5):266–271. https://doi.org/10.1007/s12250-013-3340-7
Yoshida T, Nagasaki K, Takashima Y, Shirai Y, Tomaru Y, Takao Y, Sakamoto S, Hiroishi S, Ogata H (2008) Ma-LMM01 infecting toxic Microcystis aeruginosa illuminates diverse cyanophage genome strategies. J Bacteriol 190(5):1762–1772. https://doi.org/10.1128/JB.01534-07
Yang F, Jin H, Wang XQ, Li Q, Zhang JT, Cui N, Jiang YL, Chen Y, Wu QF, Zhou CZ, Li WF (2020) Genomic analysis of Mic1 reveals a novel freshwater long-tailed cyanophage. Front Microbiol 11:484. https://doi.org/10.3389/fmicb.2020.00484
Lin W, Li D, Sun Z, Tong Y, Yan X, Wang C, Zhang X, Pei G (2020) A novel freshwater cyanophage vB_MelS-Me-ZS1 infecting bloom-forming cyanobacterium Microcystis elabens. Mol Biol Rep 47(10):7979–7989. https://doi.org/10.1007/s11033-020-05876-8
Naknaen A, Suttinun O, Surachat K, Khan E, Pomwised R (2021) A novel jumbo phage PhiMa05 inhibits harmful Microcystis sp. Front Microbiol 12:660351. https://doi.org/10.3389/fmicb.2021.660351
Qian M, Li D, Lin W, Pan L, Liu W, Zhou Q, Cai R, Wang F, Zhu J, Tong Y (2022) A novel freshwater cyanophage, Mae-Yong924-1, reveals a new family. Viruses 14(2):283. https://doi.org/10.3390/v14020283
Zhang X, Kang H, Li Y, Liu X, Yang Y, Li S, Pei G, Sun Q, Shu P, Mi Z, Huang Y, Zhang Z, Liu Y, An X, Xu X, Tong Y (2015) Conserved termini and adjacent variable region of Twortlikevirus Staphylococcus phages. Virol Sin 30(6):433–440. https://doi.org/10.1007/s12250-015-3643-y
Garneau JR, Depardieu F, Fortier LC, Bikard D, Monot M (2017) PhageTerm: a tool for fast and accurate determination of phage termini and packaging mechanism using next-generation sequencing data. Sci Rep 7(1):8292. https://doi.org/10.1038/s41598-017-07910-5
Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R (2014) The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res 42(Database issue):D206–D214. https://doi.org/10.1093/nar/gkt1226
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215(3):403–410. https://doi.org/10.1016/S0022-2836(05)80360-2
Potter SC, Luciani A, Eddy SR, Park Y, Lopez R, Finn RD (2018) HMMER web server: 2018 update. Nucleic Acids Res 46(W1):W200–W204. https://doi.org/10.1093/nar/gky448
Zimmermann L, Stephens A, Nam SZ, Rau D, Kübler J, Lozajic M, Gabler F, Söding J, Lupas AN, Alva V (2018) A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J Mol Biol 430(15):2237–2243. https://doi.org/10.1016/j.jmb.2017.12.007
Schattner P, Brooks AN, Lowe TM (2005) The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res 33(Web Server issue):W686–W689. https://doi.org/10.1093/nar/gki366
Lee I, Ouk Kim Y, Park SC, Chun J (2016) OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol 66(2):1100–1103. https://doi.org/10.1099/ijsem.0.000760
Bao Y, Chetvernin V, Tatusova T (2014) Improvements to pairwise sequence comparison (PASC): a genome-based web tool for virus classification. Adv Virol 159(12):3293–3304. https://doi.org/10.1007/s00705-014-2197-x
Moraru C, Varsani A, Kropinski AM (2020) VIRIDIC-a novel tool to calculate the intergenomic similarities of prokaryote-infecting viruses. Viruses 12(11):1268. https://doi.org/10.3390/v12111268
De Jonge PA, Nobrega FL, Brouns S, Dutilh BE (2019) Molecular and evolutionary determinants of bacteriophage host range. Trends Microbiol 27(1):51–63. https://doi.org/10.1016/j.tim.2018.08.006
Hu NT, Thiel T, Giddings TH, Jr Wolk CP (1981) New Anabaena and Nostoc cyanophages from sewage settling ponds. Virology 114(1):236–246. https://doi.org/10.1016/0042-6822(81)90269-5
De Leeuw M, Baron M, Ben David O, Kushmaro A (2020) Molecular insights into bacteriophage evolution toward its host. Viruses 12(10):1132. https://doi.org/10.3390/v12101132
Mittler JE (1996) Evolution of the genetic switch in temperate bacteriophage. I. Basic theory. J Theor Biol 179(2):161–172. https://doi.org/10.1006/jtbi.1996.0056
Suttle CA, Chan AM (1994) Dynamics and distribution of cyanophages and their effect on marine Synechococcus spp. Appl Environ Microbiol 60(9):3167–3174. https://doi.org/10.1128/aem.60.9.3167-3174.1994
Cores de Vries G, Wu XS, Haggård-Ljungquist E (1991) Genetic analysis of the DNA recognition sequence of the P2 Cox protein. J Virol 65(9):4665–4669. https://doi.org/10.1128/JVI.65.9.4665-4669.1991
Flodman K, Tsai R, Xu MY, Corrêa IR, Jr Copelas A, Lee YJ, Xu MQ, Weigele P, Xu SY (2019) Type II restriction of bacteriophage DNA with 5hmdU-derived base modifications. Front Microbiol 10:584. https://doi.org/10.3389/fmicb.2019.00584
Wion D, Casadesús J (2006) N6-methyl-adenine: an epigenetic signal for DNA-protein interactions. Nat Rev Microbiol 4(3):183–192. https://doi.org/10.1038/nrmicro1350
Carter MQ, Pham A, Huynh S, Parker CT, Miller A, He X, Hu B, Chain P (2021) DNA adenine methylase, not the PstI restriction-modification system, regulates virulence gene expression in Shiga toxin-producing Escherichia coli. Food Microbiol 96:103722. https://doi.org/10.1016/j.fm.2020.103722
Abbes C, Joseleau-Petit D, Liébart JC, D’Ari R, Sezonov G (2001) The GemA protein of phage Mu and the GyrB gyrase subunit of Escherichia coli: the search for targets and interactions leading to the reversion of Mu-induced mutations. Biochimie 83(2):261–267. https://doi.org/10.1016/s0300-9084(00)01214-1
Kumaraswami M, Howe MM, Park HW (2004) Crystal structure of the Mor protein of bacteriophage Mu, a member of the Mor/C family of transcription activators. J Biol Chem 279(16):16581–16590. https://doi.org/10.1074/jbc.M313555200
Wippel K, Long SR (2016) Contributions of Sinorhizobium meliloti transcriptional regulator DksA to bacterial growth and efficient symbiosis with Medicago sativa. J Bacteriol 198(9):1374–1383. https://doi.org/10.1128/JB.00013-16
Nishimura Y, Yoshida T, Kuronishi M, Uehara H, Ogata H, Goto S (2017) ViPTree: the viral proteomic tree server. Bioinformatics (Oxford, Engl) 33(15):2379–2380. https://doi.org/10.1093/bioinformatics/btx157
Sullivan MJ, Petty NK, Beatson SA (2011) Easyfig: a genome comparison visualizer. Bioinformatics (Oxford, Engl) 27(7):1009–1010. https://doi.org/10.1093/bioinformatics/btr039
Acknowledgements
We are grateful for the high-quality technical support provided by Pingping Zhan of the Electron Microscopy Laboratory of Ningbo University.
Funding
This study was funded by the National Key Research and Development Program (2018YFA0903000) and the Open Fund of Key Laboratory of Biogenetic Resources of the State Oceanic Administration (HY201602) and sponsored by the K. C. Wong Magna Fund of Ningbo University.
Author information
Authors and Affiliations
Contributions
DL, RC, and YT designed the research. RC, DL, WL, WQ, LP, FW, MQ, WL, QZ, CZ, and YT performed the research. RC and DL analyzed data and wrote the paper. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Handling Editor: Johannes Wittmann.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary Fig. S1
(a) Transparent circular plaques formed by cyanophage Mwe-Yong1112-1 on an M. wesenbergii lawn. (b) Macrograph of normal FACHB-1112 cultures (1112 C) and FACHB-1112 co-inoculated with cyanophage Mwe-Yong1112-1 (1112 T). Supplementary file1 (TIF 23201 KB)
Rights and permissions
About this article
Cite this article
Cai, R., Li, D., Lin, W. et al. Genome sequence of the novel freshwater Microcystis cyanophage Mwe-Yong1112-1. Arch Virol 167, 2371–2376 (2022). https://doi.org/10.1007/s00705-022-05542-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-022-05542-3