Abstract
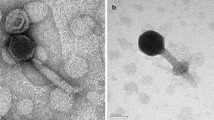
Cyanophages, which play a significant role in food web and global biochemical cycle, are one of the main causes of microbial death in aquatic environment. A novel cyanophage S-B68 was isolated from the surface water of the Bohai Sea, northern China. It can infect marine Synechococcus sp. (strain WH7803). The transmission electron microscopy results demonstrate that this cyanophage has an icosahedral head (51 nm in diameter) and a long tail (110 nm in length) and belongs to family Siphophages. The complete genome sequence of cyanophage S-B68 contains a linear, double-stranded 163,982 bp DNA molecule with a 51.7% G+C content. Except for four tRNAs, the genome contains 229 open reading frames (ORFs) which were grouped into six functional modules as follows: structure, hypothetical protein, DNA replication and expression, lysis, packaging, and some additional functions. It was found in one-step growth curve that the latent period of the S-B68 was about 49 h after infection with Synechococcus, and then it entered the rising period, and tended to stable after 61 h. Using the BLASTN tool in the NCBI database for genome comparison, there was no significant similarity between S-B68 and other known cyanophages. Present study adds a novel Siphoviridae genome to marine cyanophage dataset and provides useful basic information for its further research.



Similar content being viewed by others
References
Jassim SAA, Limoges RG (2013) Impact of external forces on cyanophage–host interactions in aquatic ecosystems. World J Microb Biot 29(10):1751–1762
Ma Y, Jiao N (2004) Molecular ecology studies of marine Synechococcus. Prog Nat Sci 14(8):649–655
Murphy LS, Haugen EM (1985) The distribution and abundance of phototrophic ultraplankton in the North Atlantic 1, 2. Limnol Oceanogr 30(1):47–58
Iturriaga R, Mitchell BG (1986) Chroococcoid cyanobacteria: a significant component in the food web dynamics of the open ocean. Mar Ecol Prog Ser 28(3):291–297
Zhong Y, Chen F, Wilhelm SW, Poorvin L, Hodson RE (2002) Phylogenetic diversity of marine cyanophage isolates and natural virus communities as revealed by sequences of viral capsid assembly protein gene g20. Appl Environ Microbiol 68(4):1576–1584
Gao EB, Huang Y, Ning D (2016) Metabolic genes within cyanophage genomes: implications for diversity and evolution. Genes 7(10):80
Weigele PR, Pope WH, Pedulla ML, Houtz JM, Smith AL, Conway JF et al (2007) Genomic and structural analysis of Syn9, a cyanophage infecting marine Prochlorococcus and Synechococcus. Environ Microbiol 9(7):1675–1695
Børsheim KY, Bratbak G, Heldal M (1990) Enumeration and biomass estimation of planktonic bacteria and viruses by transmission electron microscopy. Appl Environ Microbiol 56(2):352–356
Chen F, Lu J (2002) Genomic sequence and evolution of marine cyanophage P60: a new insight on lytic and lysogenic phages. Appl Environ Microbiol 68(5):2589–2594
Suttle CA, Chan AM, Cottrell MT (1990) Infection of phytoplankton by viruses and reduction of primary productivity. Nature 347(6292):467
Wommack KE, Colwell RR (2000) Virioplankton: viruses in aquatic ecosystems. Microbiol Mol Biol Rev 64(1):69–114
Liu Z, Wang M, Meng X, Li Y, Wang D, Jiang Y et al (2017) Isolation and genome sequencing of a novel Pseudoalteromonas phage PH1. Curr Microbiol 74(2):212–218
Sun M, Lin H, Wang M, Liu Y, Liu Z, Meng X et al (2018) Characterization and complete genome of the marine Pseudoalteromonas phage PH103, isolated from the Yellow Sea, China. Mar Genom 42:67–70
Zhu M, Wang M, Jiang Y, You S, Zhao G, Liu Y et al (2018) Isolation and complete genome sequence of a novel Marinobacter phage B23. Curr Microbiol 75(12):1619–1625
Brussaard CPD (2004) Optimization of procedures for counting viruses by flow cytometry. Appl Environ Microbiol 70(3):1506–1513
You S, Wang M, Jiang Y, Jiang T, Liu Y, Liu X et al (2019) The genome sequence of a novel cyanophage s-b64 from the Yellow Sea, China. Curr Microbiol 76:781–786
Li J, Liu W, Gao S, Warren A, Song W (2013) Multigene-based analyses of the phylogenetic evolution of oligotrich ciliates, with consideration of the internal transcribed spacer 2 secondary structure of three systematically ambiguous genera. Eukaryot Cell 12(3):430–437
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32(5):1792–1797
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41:95–98
Gouy M, Guindon S, Gascuel O (2009) SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27(2):221–224
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24(8):1596–1599
Meng X, Wang M, You S, Wang D, Li Y, Liu Z et al (2017) Characterization and complete genome sequence of a novel Siphoviridae bacteriophage BS5. Curr Microbiol 74(7):815–820
Gong Z, Wang M, Yang Q, Li Z, Xia J, Gao Y et al (2017) Isolation and complete genome sequence of a novel Pseudoalteromonas phage PH357 from the Yangtze River Estuary. Curr Microbiol 74(7):832–839
Fokine A, Chipman PR, Leiman PG, Mesyanzhinov VV, Rao VB, Rossmann MG (2004) Molecular architecture of the prolate head of bacteriophage T4. Proc Natl Acad Sci USA 101(16):6003–6008
Miller ES, Kutter E, Mosig G, Arisaka F, Kunisawa T, Rüger W (2003) Bacteriophage T4 genome. Microbiol Mol Biol Rev 67(1):199–201
Acknowledgements
We are grateful to the Marine S&T Fund of Shandong Province for Pilot National Laboratory for Marine Science and Technology (Qingdao) (Nos. 2018SDKJ0406-6 and 2018SDKJ0104-4), the National Key Research and Development Program of China (Nos. 2018YFC1406704 and 2017YFA0603200), the National Natural Science Foundation of China (Nos. 31500339 and 41076088), and Fundamental Research Funds from the Central University of Ocean University of China (Nos. 201812002, 201762017 and 201562018). We appreciate the captain and crews of the RV ‘Dongfanghong 2′ and thank Prof. Hongbin Liu for providing the synechococcus host. The authors would like to thank the editor and anonymous reviewers for their constructive comments.
Author information
Authors and Affiliations
Contributions
YJ participated in experimental design; LH, QL, XL and QZ were involved in the discussion; LH performed the main experiments and drafted the manuscript; MW, HS, CG, YL and YJ revised the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Huang, L., Liu, Q., Liu, X. et al. Isolation and Complete Genome Sequence of a Novel Cyanophage S-B68. Curr Microbiol 77, 2385–2390 (2020). https://doi.org/10.1007/s00284-020-02045-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-020-02045-9