Abstract
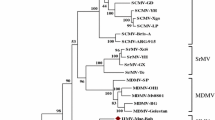
Cereal chlorotic mottle virus (CCMoV) is a cicadellid-transmitted plant rhabdovirus associated with chlorotic and necrotic streaks on several gramineous hosts and weeds. The virus was initially described in 1979 in Australia, but its genome has never been sequenced. In this study, the complete genome sequence of a Moroccan isolate of CCMoV was generated by high-throughput sequencing from infected oat leaves (Avena sativa). The genome is 13,800 nt long, containing seven open reading frames (ORFs) arranged in the canonical organization of rhabdoviruses: 3’-nucleocapsid (N), phosphoprotein (P), unknown protein (p3), unknown protein (p4), matrix (M), glycoprotein (G), viral polymerase (L)-5’. Pairwise analysis showed that maize fine streak virus (MFSV, genus Gammanucleorhabdovirus) was the closest relative. The amino acid identity values between homologous proteins from CCMoV and MFSV are as follows: 59.27% (N), 36.7% (P), 24% (P3), 62% (P4), 43.70% (M), 49.15% (G), 60.93% (L). Based on its phylogenetic relationship and analogous genome architecture, CCMoV should be assigned as member of the genus Gammanucleorhabdovirus. The low sequence similarity observed between CCMoV and MFSV suggests that CCMoV is a member of a distinct virus species.


Similar content being viewed by others
Data availability
The genomic sequence determined in this study was submitted to the GenBank database under the accession number MW731536.
References
Walker PJ, Freitas-Astúa J, Bejerman N et al (2021) ICTV virus taxonomy profile: Rhabdoviridae. J Gen Virol (in press)
Greber RS (1979) Cereal chlorotic mottle virus—a rhabdovirus of Gramineae in Australia transmitted by Nesoclutha pallida (Evans). Aust J Agric Res 30:433–443. https://doi.org/10.1071/AR9790433
Lockhart BEL (1986) Ocurrence of cereal chlorotic mottle virus in Northern Africa. Plant Dis 70:912–915. https://doi.org/10.1094/PD-70-912
Greber RS, Gowanfock DH (1979) Cereal chlorotic mottle virus purification, serology and electron microscopy in plant and insect tissues. Aust J Biol Sci 32:399–408. https://doi.org/10.1071/BI9790399
Kearse M, Moir R, Wilson A et al (2012) Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. https://doi.org/10.1093/bioinformatics/bts199
Bushnell B (2022) BBTools software package. https://sourceforge.net/projects/bbmap/
Krogh A, Larsson B, Von Heijne G, Sonnhammer ELL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580. https://doi.org/10.1006/jmbi.2000.4315
Nguyen Ba AN, Pogoutse A, Provart N, Moses AM (2009) NLStradamus: a simple Hidden Markov Model for nuclear localization signal prediction. BMC Bioinform 10:1–11. https://doi.org/10.1186/1471-2105-10-202
Tsai C, Redinbaugh MG, Willie KJ et al (2005) Complete genome sequence and in planta subcellular localization of maize fine streak virus proteins. J Virol 79:5304–5314. https://doi.org/10.1128/JVI.79.9.5304
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. https://doi.org/10.1093/molbev/mst010
Suchard MA, Lemey P, Baele G et al (2018) Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol 4:1–5. https://doi.org/10.1093/ve/vey016
Acknowledgements
Robert A. Alvarez-Quinto is supported by the Republic of Ecuador through the Ecuadorean Science and Technology Secretariat (SENESCYT).
Funding
This work was supported by the USDA National Institute of Food and Agriculture, Hatch project #1009997 and the University of Minnesota, Twin Cities.
Author information
Authors and Affiliations
Contributions
Conceptualization: Benham E. Lockhart, Dimitre Mollov. Formal analysis: Robert A. Alvarez-Quinto, Samuel Grinstead. Investigation: Robert A. Alvarez-Quinto, Benham E. Lockhart, Joana Serrano. Methodology: Benham E. Lockhart, Dimitre Mollov. Supervision: Benham E. Lockhart. Writing—original draft: Robert A. Alvarez-Quinto. Writing—review & editing: Robert A. Alvarez-Quinto, Benham E. Lockhart, Joana Serrano, Samuel Grinstead, Dimitre Mollov. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Research involving human participants and/or animals
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Handling Editor: Ralf Georg Dietzgen.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.

705_2022_5527_MOESM1_ESM.jpg
Supplementary file1 Supplementary Fig. S1 Sequence alignment of the gene junctions detected in cereal chlorotic mottle virus (CCMoV-M) and maize fine streak virus (MFSV). All sequence differences are highlighted in black (JPG 69 KB)
Rights and permissions
About this article
Cite this article
Alvarez-Quinto, R.A., Lockhart, B.E., Serrano, J. et al. Complete genome sequence of a Moroccan isolate of cereal chlorotic mottle virus. Arch Virol 167, 2347–2350 (2022). https://doi.org/10.1007/s00705-022-05527-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-022-05527-2