Abstract
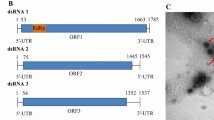
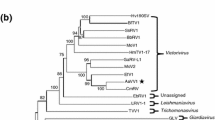
In the present study, a novel double-stranded RNA (dsRNA) mycovirus designated as "Alternaria tenuissima partitivirus 2" (AttPV2), was isolated from Alternaria tenuissima strain BJ-SY-1, a phytopathogenic fungus causing muskmelon leaf blight in Beijing municipality of China. The genome of AttPV2 comprises two dsRNA segments. The larger segment is 1829 bp in length and has a single open reading frame (ORF), potentially encoding a 65.8-kDa RNA-dependent RNA polymerase (RdRp), and the smaller segment is 1681 bp in length and also contains a single ORF, encoding a putative coat protein (CP) with a molecular mass of 56.0 kDa. BLASTp analysis revealed that the RdRp and CP encoded by the two ORFs of AttPV2 have the highest sequence identity, 85.9% and 75.0%, respectively, to their counterparts in Colletotrichum eremochloae partitivirus 1 (CePV1). Phylogenetic analysis based on RdRp sequences showed that AttPV2 clustered most closely with CePV1, a member of the proposed genus “Epsilonpartitivirus” in the family Partitiviridae. Hence, we propose that AttPV2 is a new epsilonpartitivirus from A. tenuissima. To the best of our knowledge, this is the first report of an epsilonpartitivirus infecting A. tenuissima.


Similar content being viewed by others
References
Pearson MN, Beever RE, Boine B, Arthur K (2009) Mycoviruses of filamentous fungi and their relevance to plant pathology. Mol Plant Pathol 10:115–128
Ghabrial SA, Suzuki N (2009) Viruses of plant pathogenic fungi. Annu Rev Phytopathol 47:353–384
Ghabrial SA, Castón JR, Jiang DH, Nibert ML, Suzuki N (2015) 50-plus years of fungal viruses. Virology 479–480:356–368
Wan XR, Zhao Y, Zhang YY, Wei CX, Du HY, Zhang H, Chen JH, Yang LJ, Zang R, Wen CY (2021) Molecular characterization of a novel partitivirus isolated from the phytopathogenic fungus Aplosporella javeedii. Arch Virol 166:1237–1240
Nibert ML, Tang JH, Xie JT, Collier AM, Ghabrial SA, Baker TS, Tao YJ (2013) 3D structures of fungal partitiviruses. Adv Virus Res 86:59–85
Vainio EJ, Chiba S, Ghabrial SA, Maiss E, Roossinck M, Sabanadzovic S, Suzuki N, Xie JT, Nibert ML, Consortium IR (2018) ICTV virus taxonomy profile: Partitiviridae. J Gen Virol 99:17–18
Nerva L, Silvestri A, Ciuffo M, Palmano S, Varese GC, Turina M (2017) Transmission of Penicillium aurantiogriseum partiti-like virus 1 to a new fungal host (Cryphonectria parasitica) confers higher resistance to salinity and reveals adaptive genomic changes. Environ Microbiol 19:4480–4492
Jiang YH, Wang JX, Yang B, Wang QR, Zhou JJ, Yu WF (2019) Molecular characterization of a debilitation-associated partitivirus infecting the pathogenic fungus Aspergillus flavus. Front Microbiol 10:626
Gilbert KB, Holcomb EE, Allscheid RL, Carrington JC (2019) Hiding in plain sight: new virus genomes discovered via a systematic analysis of fungal public transcriptomes. PLoS ONE 14:e0219207
Shi NJ, Yang GG, Wang P, Wang YL, Yu DS, Huang B (2019) Complete genome sequence of a novel partitivirus from the entomogenous fungus Beauveria bassiana in China. Arch Virol 164:3141–3144
Wang P, Yang GG, Shi NJ, Huang B (2020) Molecular characterization of a new partitivirus, MbPV1, isolated from the entomopathogenic fungus Metarhizium brunneum in China. Arch Virol 165:765–769
Zhu JZ, Guo J, Hu Z, Zhang XT, Li XG, Zhong J (2021) A novel partitivirus that confer hypovirulence to the plant pathogenic fungus Colletotrichum liriopes. Front Microbiol 12:653809
Shi M, Lin XD, Tian JH, Chen LJ, Chen X, Li CX, Qin XC, Li J, Cao JP, Eden JS, Buchmann J, Wang W, Xu JG, Holmes EC, Zhang YZ (2016) Redefining the invertebrate RNA virosphere. Nature 540:539–543
Woudenberg JHC, Groenewald JZ, Binder M, Crous PW (2013) Alternaria redefined. Stud Mycol 75:171–212
Thomma BP (2003) Alternaria spp.: from general saprophyte to specific parasite. Mol Plant Pathol 4:225–236
Ma GP, Liang ZJ, Hua HH, Zhou T, Wu XH (2019) Complete genome sequence of a new botybirnavirus isolated from a phytopathogenic Alternaria alternata in China. Arch Virol 164:1225–1228
Ma GP, Zhang XF, Hua HH, Zhou T, Wu XH (2020) Molecular and biological characterization of a novel strain of Alternaria alternata chrysovirus 1 identified from the pathogen Alternaria tenuissima causing watermelon leaf blight. Virus Res 280:197904
Liang ZJ, Wang XY, Hua HH, Cao W, Zhou T, Zhao C, Wu XH (2021) Full genome sequence of a new three-segment gammapartitivirus from the phytopathogenic fungus Alternaria tenuissima on cotton in China. Arch Virol 166:973–976
Zheng HH, Zhao J, Wang TY, Wu XH (2015) Characterization of Alternaria species associated with potato foliar diseases in China. Plant Pathol 64:425–433
Morris TJ, Dodds JA (1979) Isolation and analysis of double-stranded RNA from virus-infected plant and fungal tissue. Phytopathology 69:854–858
Darissa O, Willingmann P, Adam G (2010) Optimized approaches for the sequence determination of double-stranded RNA templates. J Virol Methods 169:397–403
Li SW, Li YT, Hu CH, Han CG, Zhou T, Zhao C, Wu XH (2020) Full genome sequence of a new mitovirus from the phytopathogenic fungus Rhizoctonia solani. Arch Virol 165:1719–1723
Lau SKP, Lo GCS, Chow FWN, Fan RYY, Cai JJ, Yuen KY, Woo PCY (2018) Novel partitivirus enhances virulence of and causes aberrant gene expression in Talaromyces marneffei. MBio 9:e00947-e1018
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Xiang J, Fu M, Hong N, Zhai LF, Xiao F, Wang GP (2017) Characterization of a novel botybirnavirus isolated from a phytopathogenic Alternaria fungus. Arch Virol 162:3907–3911
Funding
This work was financially supported by Beijing Innovation Consortium of Melon Research System (Grant no. BAIC10-2021).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This study did not include experiments with human participants or animals performed by any of the authors.
Additional information
Handling Editor: Massimo Turina.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary Fig. S1
Multiple sequence alignments of the 5′ and 3′ untranslated regions (UTRs) of the coding strands of the two double-stranded RNAs (dsRNAs) of Alternaria tenuissima partitivirus 2 (AttPV2). Black shading indicates identical nucleotides at both ends. (TIF 1169 KB)
Rights and permissions
About this article
Cite this article
Wang, X., Ma, Z., Li, R. et al. Complete genome sequence of a novel partitivirus infecting the phytopathogenic fungus Alternaria tenuissima. Arch Virol 167, 635–639 (2022). https://doi.org/10.1007/s00705-021-05332-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-021-05332-3