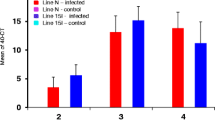
Abstract
Infectious bronchitis virus (IBV) is one of the major threats to the poultry industry, with significant economic consequences. Despite strict measures, the disease is difficult to control worldwide. Experimental evidence demonstrates that the severity of IBV is affected by the genetic background of the chicken, and the selection of appropriate breeds can increase production efficiency. Therefore, the aim of the present study was to assess the strength of the immune response to IBV in tracheal tissues of Ross 308 and Cobb 500 broiler chickens by evaluating transcriptome changes, focusing on immune responses and the viral load in tracheal tissues two days after IBV infection. We identified 899 and 1350 differentially expressed genes (DEGs) in the Cobb 500 and Ross 308 experimental groups compared to their respective control groups. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis indicated the involvement of signaling pathways (Toll-like receptor [TLR], NOD-like receptor [NLR], and RIG-I-like receptor [RLR] signaling pathways). Interestingly, the RLR signaling pathway appears to be affected only in the Cobb hybrid. Furthermore, the viral loads in tracheal samples obtained from the Ross challenged group were significantly higher than those of the Cobb challenged group. The results of this study indicated that the host transcriptional response to IBV infection as well as the viral load can differ by hybrid. Furthermore, genes such as TLR-3, ChIFN-α, MDA5, LGP2, IRF-7, NF-κB, and TRIM25 may interfere with IBV proliferation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
With the ever-growing human population, there is a strong tendency for poultry to become the main protein source. Therefore, the poultry industry plays an important role in the food supply chain, and its economic significance cannot be ignored [1]. However, this industry faces various challenges, among which are infectious diseases, and viral diseases can cause huge economic losses to the industry worldwide [1, 2]. Infectious bronchitis virus (IBV) is one of the major threats to the industry and has significant economic consequences [1]. The causative agent of IBV is a positive-sense single-stranded RNA virus of the family Coronaviridae [3, 4].
The virus is known for its rapid reproduction with high mutation and recombination frequency, leading to geographically distinct strains [1, 5]. The virus enters through and replicates in the host’s respiratory tract before respiratory signs start to appear within 24 to 48 hours of contact with the virus, and this is known as the incubation period [6, 7]. Some IBV strains may have tropism for other epithelial tissues (including the kidneys and oviduct), in which they cause infection and lesions [8,9,10]. The disease can be exacerbated when it coincides with secondary bacterial infections.
IBV affects chickens of all ages and can be transmitted rapidly across farms. Within three days of infection, clinical signs such as depression, poor weight gain, and appetite loss may appear. Furthermore, lesions caused by some strains in the oviduct markedly decrease egg quality and production [10, 11]. In addition, the mortality rate can be as high as 83%, depending on the virus strain and health condition of the chickens [7, 12]. Considering the above facts, the poultry industry must adopt and implement policies and preemptive measures. Current preventive strategies are a hybrid of appropriate management practices, including temperature control mechanisms and advanced ventilation, and the use of attenuated and inactivated viral vaccines [13, 14]. Despite these measures, however, the disease has proven difficult to control worldwide due to the diversity of strains in different geographical regions and the absence of complete cross-protection between them [15]. Hence, developing preventive and therapeutic treatments is necessary to control the spread of the virus. Previous studies have suggested several ways to combat the disease. For instance, Jackwood et al. suggested that polyvalent vaccines containing circulating virus strains can be used for protection against the virus [5]. Moreover, recombinant vaccines containing S1 and N proteins have been reported to induce an immune response [16].
Although numerous studies have been conducted, the control of IBV continues to be a major challenge. To introduce more-effective vaccines and develop enhanced preventive measures, it is essential to understand defense mechanisms against poultry pathogens. Such understanding can provide invaluable insights into molecular pathogenesis in birds and various pathways involved in virus-host interactions. Recent advances in genomics, transcriptomics, proteomics, and metabolomics are now often incorporated into methodologies to analyze cellular and molecular interactions in biological systems [17].
The use of high-throughput techniques, such as microarray and next-generation sequencing (NGS), has gained momentum among researchers exploring the interactions between IBV and the host [3, 13, 14, 17,18,19]. These investigations have provided intriguing insights into how antiviral mechanisms in poultry function against the virus causing infectious bronchitis. To the best of our knowledge, no study has looked into transcriptome changes in the Ross 308 and Cobb 500 hybrids to investigate their antiviral responses to IBV.
Previous studies have revealed that the viral load can differ in different tissues of chickens infected with IBV [20, 21]. However, there is limited information about the evaluation of viral load in different hybrids of chickens infected with IBV. Additionally, the selection of appropriate genetic breeds has been shown to increase production efficiency [22]. Therefore, the aim of the present study was to evaluate transcriptome changes (by focusing on immune responses) and viral load in tracheal tissues of Ross 308 and Cobb 500 broiler chickens’ infected with IBV using RNA sequencing (RNA-seq) and quantitative reverse transcription polymerase chain reaction (RT-qPCR) methods.
Materials and methods
Experimental design
In this randomized controlled experimental study, a total of 120 one-day-old female broiler chickens (Ross 308 and Cobb 500) were randomly divided into four experimental and control groups (n = 30): Cobb control (Cobb Con), Ross control (Ross Con), Cobb experimental (Cobb Exper), and Ross experimental (Ross Exper). The two hybrids were kept in separate rooms (60 chickens per room). The chickens were fed and watered separately in subgroups of 15. To avoid interference by maternal antibodies, an IDEXX IBV Ab ELISA Kit (USA) was used to measure anti-IBV antibody titers after three weeks according to the manufacturer’s instructions. Chickens that were negative for anti-IBV antibodies were assigned to experimental subgroups, and chickens with positive antibody test results were excluded from the study. Immediately after this procedure, chickens in the experimental groups were challenged intranasally with 200 μL of allantoic fluid containing 104 times the 50% egg infective dose (EID50)/mL of the Var2-like (IS/1494/06-like) strain. Control groups received 0.2 mL of sterile phosphate-buffered saline (PBS) intranasally.
Virus strain and titration
The Iranian variant-2-like IBV strain IS/1494 was used in the study. The strain was isolated from an Iranian broiler farm in 2014 [23]. The EID50 was calculated using the Reed–Muench method [24].
Sample collection and preparation
Two days after challenge, 10 chickens were randomly selected from each subgroup and euthanized according to the University of Tehran’s ethical principles (i.e., inhalation of carbon dioxide followed by dislocation of their cervical vertebrae) under sterile conditions. Then, tracheal tissues were dissected and pooled for each subgroup and placed in sterile 5-mL tubes containing RNAlater solution (Thermo Fisher Scientific, Massachusetts, USA) to stabilize and protect RNA with immediate RNase inactivation. In addition, tissue was removed from individual chickens in each subgroup and placed in sterile 1.5-mL tubes containing RNAlater solution to measure the viral load and perform a RT-qPCR test to evaluate RNA-seq results. The pooled tracheal tissues were sent to BGI in China for paired-end sequencing using a HiSeq 2000 system and a TruSeq SBS Kit v3-HS Illumina. All RNA extraction processes and quality control were performed by the company.
Total RNA from all pooled samples was extracted using the TRIzol method according to the instructions of the TRIzol Kit Manual (B518651-0100, Sangon Biotech, Shanghai, China). The RNA purity was determined based on spectroscopic measurements at 230, 260, and 280 nm using a spectrophotometer (IMPLEN, CA, USA), and the integrity of the RNA was assessed using a Bioanalyzer 2100 system and an RNA Nano Assay Kit (Agilent Technologies, Santa Clara, CA, USA). The RNA samples that passed the purity and integrity tests (i.e., RNAs with a minimum RNA integrity number (RIN) of 8.00, a A260/280 ratio of at least 1.80, and an A260/230 ratio of at least 2.00) were used for transcriptome sequencing using an Illumina HiSeq 2000 platform.
Transcriptome analysis
About 31 million paired-end raw reads for each sample with a length of 90–100 base pairs (bp) were generated from RNA-seq libraries in FASTQ format. Before read alignment, the FastQC tool (https://www.bioinformatics.babraham.ac.uk/projects/fastqc) was used for quality control of raw reads, and the reads were pre-filtered to remove probable contaminants. Quality trimming increased the number of good reads and abundance measurement accuracy. Adapter sequences and low-quality nucleotide calls in the raw data were removed using BBDuk 38.87 (BBtools, California, USA). For adapter sequence trimming, we used the following parameters: ktrim = r, k = 23, mink = 11, and hdist = 1. For nucleotide trimming, the nucleotide calls with a quality score of 20 or higher were considered high-quality reads. We defined r and l (right and left side trimming) for the “--qtrim = rl” parameter and passed quality score 20 for quality trimming thresholds to the “--trimq” parameter.
After trimming, to identify reads originating from the chicken genome, we aligned clean reads to the reference genome sequence GRCg6a (GCA_000002315.5) using Spliced Transcripts Alignment to a Reference (STAR) with default settings.
The reference genome sequence and annotation files were obtained from the Ensembl database (https://asia.ensembl.org/info/data/ftp/index.html), and genome indexing was done using the “--runMode genomeGenerate” option in the STAR software package 2.7. [25]. After alignment, several files were created, and the “ReadsPerGene.out.tab” file (a tab-delimited text file with read count per gene used for expression analysis) was the main file used for subsequent analysis to identify differentially expressed genes (DEGs).
Identification of DEGs
DEGs were identified using the edgeR software package version 3.28.1. Here, only genes with a log fold change ≥1 or ≥-1 (P < 0.05) were identified as DEGs. To create the figure showing DEGs, we constructed a heat map using the R software package (pheatmap) version 1.0.12 (https://cloud.r-project.org/web/packages/pheatmap). An MD plot was made to show the log-fold change of DEGs in the experimental groups compared to the controls, and the average abundance of each gene was determined using the R software package (plotMD).
Functional annotation analysis
To identify biological functions related to DEGs, the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways and Gene Ontology (GO) terms (cellular component, molecular function, and biological process) were investigated using the DAVID database (version 6.8) (https://david.ncifcrf.gov/) [26]. GO terms and KEGG pathways with a P-value < 0.05 were considered to exhibit statistically significant enrichment.
Validation of RNA-seq results and calculation of viral load using RT-qPCR
An RT-qPCR assay was used to evaluate and confirm the results obtained using the RNA-seq method and to calculate the viral load, in individual Cobb and Ross chickens in the study groups.
Total RNA was extracted from individual samples taken from each subgroup using a SinaPure RNA Kit (SinaClon, Iran) and treated with DNase I. The purity of the total RNA was determined using spectroscopic measurements at 230, 260, and 280 nm. The RNA integrity was evaluated by electrophoresis on a 1% agarose gel after staining with ethidium bromide and visualization under UV light (for more details, see Supplementary File 1). RNA samples with an OD260/280 ratio of 1.9–2.2 and an OD260/230 ratio greater than 2 and with high integrity (i.e., RNAs with clearly visible bands of 18S and 28S ribosomal RNA) were used for cDNA synthesis.
cDNA synthesis was performed using a RevertAid First Strand cDNA Synthesis Kit (Fermentas, Canada) as follows: a mixture of 2 μg of total RNA (7 μL RNA), 1 μL of random hexamer primer, and 4 μL of DEPC-treated water was heated at 65 °C for 10 min and cooled on ice. Then, 4 μL of 5X reaction buffer, 1 μL of Ribolock Rnase inhibitor (20 U/μL), 2 μL of 10 mM dNTP mix, and 1 μL of RevertAid M-MuLV Reverse Transcriptase (200 U/μL) were added to the solution to a final volume of 20 μL.
To validate RNA-seq results, 22 genes were selected randomly from each up- and downregulated DEG from both hybrids. All RT-qPCR primers were designed using Primer-BLAST [27], and their sequences are shown in Tables 1 and 2. For RT-qPCR, the 20-μL reaction mixture contained 2 µL of cDNA, 10 µL of SYBR Premix (SinaClon, Iran), 0.4 µL of forward and 0.4 µL of reverse primers at a final concentration of 10 μm, and 7.2 µL of RNase-free water. The reaction condition were as follows: 95 °C for 5 min, followed by 40 cycles of 95 °C for 15 s, 56 °C (CDRT1, NRROS, MHM2, TLR7, THEMIS, BEST1), 55 °C (LGP2, GAPDH, PLS1, TBX21, TLR1B), 51 °C (FAM81A, AHR2, MUC13, MYH15, RALY, FCN2, SBK2), or 48 °C (MDA5, LUZP2, ARHGAP15, GPR171, ADAP2) for 20 s, and 72 °C for 15 s using a Corbett Life Science Rotor-Gene 6000 Cycler (USA). The fold changes in expression were calculated using the 2-∆∆CT method [28]. All samples used for RT-qPCR were analyzed in triplicate.
To calculate the viral load, cDNAs from individual tracheal samples in each subgroup were used to amplify a conserved sequence of the IBV genome within the 5’ untranslated region (UTR) by real-time PCR. The forward primer 5′-GCTTTTGAGCCTAGCGTT-3′, located at nucleotide position 391–408 of the genome sequence of IBV strain M41, the reverse primer 5′-GCCATGTTGTCACTGTCTATTG-3′, located at nucleotide position 512–533, and a TaqMan dual-labeled probe (IBV5'G probe 5'-FAMCACCACCAGAACCTGTCACCTC-BHQ1-3') located at nucleotide position 473–494 were designed to amplify and detect a 143-bp fragment of the 5'-UTR [20, 29]. The 20-μL reaction mixture contained 2 μL of 10x PCR buffer (Sinaclon), 1 μL of dNTP mix (Sinaclon), 0.8 μL of 50 mM MgCl2 (Sinaclon), 0.2 μL of CinnaGen Taq DNA polymerase (Sinaclon), 5 μL of template cDNA, 0.4 µL of forward and 0.4 µL of reverse primer at a concentration of 20 μm, 0.4 µL of probe at a concentration of 20 μm, and 9.8 µL of RNase-free water. The reaction was performed using a Corbett Life Science Rotor-Gene 6000 Cycler. The PCR cycling parameters were 95 °C for 3 min, followed by then 44 cycles of 95 °C for 15 s, 60 °C for 30 s, and 72 °C for 20 s [20].
Data analysis
An independent two-sample t-test was used to compare the expression levels of DEGs and the viral loads between the study groups. All analyses were performed at a significance level of 5%. Stata 14 (StataCorp, College Station, Texas, USA) was used for data analysis.
Results
Gene expression profile
The number of raw, trimmed, and mapped reads and the mapping efficiency for each sample are presented in Table 3. After filtering, the overall QC percentage of high-quality clean data was above 95%. An average of 60 million trimmed reads per sample were mapped to the reference with a mapping efficiency of 80.90%–89.00%. Of the annotated genes in the chicken genome, 899 in Cobb 500 chickens (723 upregulated and 176 downregulated [Supplementary File 2]) and 1350 in Ross 308 chickens (650 upregulated and 700 downregulated [Supplementary File 3]) were identified as DEGs when comparing the experimental groups to their respective control groups (Fig. 1). MD plot, MDS plot, and a heat map of DEGs for each hybrid is presented in Figure 2. A Venn diagram showing the overlap of DEGs between the two hybrids is presented in Figure 3.
Functional annotation of DEGs
A functional annotation analysis was performed to determine functional categories of the DEGs. The 10 most enriched GO terms in each category (biological processes, molecular function, and cellular components) related to upregulated genes in both hybrids are listed in Tables 4, 5, 6. Additionally, the five most enriched GO terms in each category related to downregulated genes in both hybrids are shown in Figure 4.
KEGG pathway analysis
KEGG pathway analysis was performed to investigate the biological functions of the DEGs [30]. The KEGG pathways were significantly enriched in the subsets of up- and downregulated genes in each hybrid. The KEGG pathway enrichment analysis showed that the upregulated genes were mainly involved in signaling pathways (Toll-like receptor [TLR] signaling, JAK-STAT signaling, NOD-like receptor [NLR] signaling, and RIG-I-like receptor [RLR] signaling pathways) (Table 7). Interestingly, the RLR signaling pathway (including IFIH1 [also known as MDA5] and DHX58 [also known as LGP2]) was affected only in the Cobb 500 hybrid. The KEGG pathway enrichment analysis results for the downregulated genes are presented in Figure 4.
Validation of RNA-seq results by RT-qPCR
qPCR analysis confirmed that the selected genes were differentially expressed between the experimental groups (Fig. 5). Furthermore, no significant difference in the expression levels of the selected DEGs was observed between the RNA-seq results and the RT-qPCR results, indicating that the RNA-seq results were indeed reliable (Tables 8 and 9). In addition, we used agarose gel electrophoresis to confirm the specificity of the real-time PCR assay (Fig. 6), and the identity of the PCR products was confirmed by Sanger sequencing.
Agarose gel electrophoresis results of DEGs selected for RT-qPCR assay. (A) Cobb. 1, PALY (155 base pair [bp]); 2, PLS1 (158 bp); 3, THEMIS (239 bp); 4, BEST1 (172 bp); 5, MUC13 (172 bp); 6, MDA5 (244 bp); 7, LGP2 (193 bp); 8, SBK2 (214 bp); 9, MHM2 (217 bp); 10, TLR1B (173 bp); 11, LUZP2 (169 bp); 12, NRROS (163 bp). (B) Ross. 1, MYH15 (163 bp); 2, TBX21 (162 bp); 3, ADAP2 (239 bp); 4, GAPDH (162 bp); 5, AHR2 (153 bp); 6, GPR171 (204 bp); 7, TLR1B (173 bp); 8, CDRT1 (175 bp); 9, FCN2 (203 bp); 10, FAM81A (188 bp); 11, ARHGAP15 (182 bp). M, 100 bp marker
Determination of the viral load by RT-qPCR
The viral load in tracheal tissues was measured two days after infection for all study groups. The virus was detected in all samples from the infected groups, and all samples from the control groups were negative. Statistical analysis showed that two days after infection, the mean viral load in the tracheal samples of the Ross challenged group was significantly higher than in the Cobb challenged group (P-value < 0.0001) (Fig. 7).
Discussion
The respiratory system is the main entry point for respiratory pathogens, including IBV. Therefore, examining changes in the transcriptome and viral load in infected respiratory tissues can provide vital details about virus-host interactions. To the best of our knowledge, this is the first study to evaluate transcriptome changes (focusing on immune responses) and viral load in tracheal tissues of Ross 308 and Cobb 500 broiler chickens infected with IBV using RNA-seq and RT-qPCR methods two days after infection.
We identified 899 genes in the Cobb 500 experimental group and 1350 genes in the Ross 308 experimental group that were expressed at different levels from those of the corresponding control group, and these results were confirmed using RT-qPCR. However, the calculated fold-change values were different. This may be due to differences in how the fold change was calculated from the RNA-seq and RT-qPCR data. Identification of DEGs provides potentially important information about the host’s responses to IBV infection.
Our analysis identified three signaling pathways, all of which are associated with innate immune responses involving pattern-recognition receptors (PRRs), including the TLR, NLR receptor, and RLR signaling pathways. Interestingly, the RLR signaling pathway was not found to respond to IBV infection in the Ross 308 hybrid. Furthermore, at two days postinfection, viral loads in the Ross tracheal samples were significantly higher than those in the Cobb tracheal samples.
The innate immune response is the first line of defense against pathogens and can be achieved through physical barriers, such as skin and mucous membranes. Other components play non-specific protective roles. These include PRRs, heterophils, macrophages, dendritic cells (DCs), natural killer (NK) cells, acute-phase proteins, the complement system, cytokines, chemokines, and other factors for immune cell trafficking and apoptosis. PRRs of the immune system, such as TLRs, RLRs, and NLRs are involved in early identification of infectious agents [31]. TLRs are key to efficient functioning because they identify protected structures in many pathogens. So far, TLR-1A, TLR-1B, TLR-2A, TLR-2B, TLR-3, TLR-4, TLR-5, TLR-7, TLR-15, and TLR-21 have been identified in chickens [32].
In this study, an increase in expression of the genes encoding TLR-1A, TLR-1B, TLR-2A, TLR-2B, TLR-4, TLR-7, and TLR-15 was found in both hybrids, although their values differed. Unlike the Ross 308 hybrid, the gene encoding TLR-3 exhibited increased expression in the Cobb 500 hybrid. TLR-4 and TLR-15 have been reported to be involved in initiating immune responses to viral infections. Their genes have been shown to undergo increased expression following infections caused by coronaviruses, including severe acute respiratory syndrome coronavirus (SARS-CoV), IBV, and mouse hepatitis virus (MHV) [33, 34]. Consistent with our study, previous studies have found increased levels of TLR-15 in the lungs of chickens infected with Marek’s disease virus (MDV), infectious bursal disease virus (IBDV), and H9N2 influenza viruses [35]. A study conducted by Wang et al. found increased expression of TLR-3 three days after infection with IBV-M41 [14]. TLR-3 has been shown to play an important role in viral immunology [14, 35, 36]. Increased levels of TLR-1A, TLR-1B, TLR-2, TLR-3, and TLR-7 have been found in the epithelial cells of 21-day-old chickens vaccinated with the attenuated IBV vaccine strain Massachusetts [37]. Kameka et al. reported an increase in TLR-3 and TLR-7 levels in tracheal and lung tissues in chickens infected with the Connecticut strain of IBV [38]. Chen et al. reported that TLR-7 mainly recognizes single-stranded RNA (ssRNA) sequences of RNA viruses that enter endosomes by endocytosis [39].
Different pathogen-associated molecular patterns (PAMPs) have been reported to activate different TLRs, inducing production of type I interferons (IFNs). Activation of IFN, in turn, induces expression of interferon-stimulated genes (ISGs) [18, 40]. Although the gene encoding IFN-α in the Cobb 500 hybrid was identified as a DEG in this study, in the Ross 308 hybrid, no increase in the expression of this gene was observed. Also, an increase in the expression of some ISGs and interferon-regulatory factors (IRFs), including IRF4, IRF5, IRF7, IRF8, IRF9, IFIT5, IFITM5, IFIH1, IFI6, IFI30, IFI35, IFI27L2, MYD88, MX1, and RSAD2 was observed in the Cobb 500 hybrid, while increased expression of IRF1, IRF5, IFIT5, IFITM5, IFI6, and IFI30 was observed in the Ross 308 hybrid.
Our results suggest that the induction of IFN-α stimulated stronger expression of ISG and IFI in the Cobb 500 hybrid than in the Ross 308 hybrid. On the other hand, it can be said that IFN-α plays a more important role in stimulating ISGs and IFIs. Furthermore, as mentioned above, the gene encoding TLR-3 exhibited increased expression in the Cobb 500 hybrid after infection. TLR-3, a member of the TLR family of proteins, recognizes viral double-stranded RNA (dsRNA) and induces type I IFN [40]. Since coronaviruses produce dsRNA intermediates during replication [17], the stronger expression of ISG and IFI observed in the Cobb 500 hybrid might be attributed to increased expression of the gene encoding TLR-3.
Phagocytic heterophils and macrophages are among the cells utilized when the innate immune system is activated [31]. It has been reported that heterophils destroy IBV-infected cells by releasing enzymes [41]. Guo et al. found increased expression of antimicrobial cytotoxic molecules, including bactericidal permeability-increasing protein (BPI) and cathepsin S (CTSS), both of which are released by degranulation of neutrophils [41]. Likewise, in the present study, both hybrids experienced increased expression of CTSS, although to different levels. Several studies have suggested that respiratory macrophages may restrict IBV proliferation in respiratory tissues [38, 41,42,43].
In line with these findings, an increase in the level of SPI-1 gene expression was observed for both hybrids. Playing a key role in macrophage and monocyte signaling pathways, SPI-1 distinguishes macrophages [44]. Increased expression of the SPI-1 gene was reported in chickens infected with IBV strain Massachusetts [37]. Increased expression was also found for a set of other immune-related genes, including complement system genes encoding C1S, C3, C1R, and C4. These results suggest that both adaptive and innate immunity play a role in the response to IBV, while the complement system acts as a bridge between the innate and adaptive immune responses to provide a comprehensive defense against pathogens [45].
Analysis of the trachea transcriptome revealed that the Cobb 500 hybrid infected with IBV also activated the RLR signaling pathway. RLRs are a group of cytosolic PRRs that trigger an innate antiviral response in infected cells. The RLR group consists of three members: RIG-I, MDA5, and LGP2. Despite the key role of RIG-I in viral infections, its gene has not been identified in chickens [46, 47].
Several studies have suggested that, in the absence of the RIG-I gene in chickens, MDA5 plays a similar role. For instance, it has been reported that influenza A virus (AIV) infection in chicken cells is detected by MDA5 [46]. Although Keestra et al. have suggested that MDA5 compensates for the lack of RIG-I in chickens [48], Barber et al. reported that the absence of RIG-I in chickens negatively affects the antiviral innate immune response induced by the RLR signaling pathway [49]. However, a study by Liniger et al. showed that MDA5 was functionally responsible for sensing AIV in the absence of RIG-I [46].
Yang et al. evaluated gene expression in chicken cells and found that IBV infection induced messenger RNA (mRNA) expression of MDA5 and LGP2 [50]. In this study, we observed an increase in the level of MDA5 and LGP2 gene expression only in the Cobb 500 hybrid. A study by Loo and Gale demonstrated that MDA5 induced the expression of the IRF7 and NF-κB genes [47]. Consistent with Loo and Gale’s study, we observed an increase in the levels of IRF7 and NF-κB only in the Cobb 500 hybrid.
The Cobb 500 hybrid also showed an increase in the level of TRIM25 gene expression. TRIM25 is a member of the tripartite motif (TRIM) family that regulates the RLR signaling pathway. The TRIM family consists of two members: TRIM25 and TRIM27. Although chickens lack a TRIM27 gene, it has been shown that TRIM25 plays a critical role in inducing antiviral responses in chickens [51]. Xu et al. have suggested that the RLR signaling pathway plays a more important role in the detection of nephropathogenic IBV infection than the TLR signaling pathway [17].
Conclusion
In this study, the Ross 308 and Cobb 500 hybrids exhibited differences in their viral loads and transcriptome profiles in tracheal tissues two days after infection with IBV. Viral loads in tracheal samples from the Ross challenged group were significantly higher than those in the Cobb challenged group. Furthermore, a larger number of downregulated genes were identified in the Ross 308 hybrid two days after IBV infection. The trachea transcriptome analysis also revealed that, unlike the Ross 308 hybrid, increased expression of TLR-3, chicken interferon α (ChIFN-α), and other components involved in the RLR signaling pathway such as MDA5, LGP2 , IRF-7, NF-κB, and TRIM25 were observed for the Cobb 500 line. TLR-3 has been reported to play an important role in the antiviral response and to induce ChIFN-α [35, 36, 40]. Furthermore, a previous study has suggested that ChIFN-α inhibits IBV replication and is associated with respiratory illness [1]. In addition, the role of the RLR signaling pathway in the innate antiviral response should not be ignored.
It appears likely that the Cobb 500 hybrid exerts a more robust immune response to IBV infection than the Ross 308 hybrid. Furthermore, the results of this study suggest that increased expression of TLR-3, ChIFN-α, and other components involved in the RLR signaling pathway (such as MDA5, LGP2, IRF-7, NF-κB, and TRIM25) may interfere with IBV proliferation. This could be considered a potential area for future research.
References
Barjesteh N, O’Dowd K, Vahedi SM (2020) Antiviral responses against chicken respiratory infections: focus on avian influenza virus and infectious bronchitis virus. Cytokine 127:154961
Nawab A et al (2019) Chicken toll-like receptors and their significance in immune response and disease resistance. Int Rev Immunol 38(6):284–306
Liu H et al (2017) Comparative transcriptome analysis reveals induction of apoptosis in chicken kidney cells associated with the virulence of nephropathogenic infectious bronchitis virus. Microb Pathog 113:451–459
Reddy VR et al (2015) Genetic characterization of the Belgian nephropathogenic infectious bronchitis virus (NIBV) reference strain B1648. Viruses 7(8):4488–4506
Jackwood MW, Hall D, Handel A (2012) Molecular evolution and emergence of avian gammacoronaviruses. Infect Genet Evol 12(6):1305–1311
Ambali A, Jones R (1990) Early pathogenesis in chicks of infection with an enterotropic strain of infectious bronchitis virus. Avian Dis 34:809–817
Awad F et al (2014) An overview of infectious bronchitis virus in chickens. Worlds Poult Sci J 70(2):375–384
Benyeda Z et al (2009) Comparison of the pathogenicity of QX-like, M41 and 793/B infectious bronchitis strains from different pathological conditions. Avian Pathol 38(6):449–456
De Wit J et al (2011) Induction of cystic oviducts and protection against early challenge with infectious bronchitis virus serotype D388 (genotype QX) by maternally derived antibodies and by early vaccination. Avian Pathol 40(5):463–471
Ganapathy K et al (2012) QX-like infectious bronchitis virus isolated from cases of proventriculitis in commercial broilers in England. Vet Rec 171(23):597
Jackwood MW, De Wit S (2013) Infectious bronchitis. Dis Poultry. https://doi.org/10.1002/9781119421481.ch4
Raj GD, Jones R (1997) Infectious bronchitis virus: immunopathogenesis of infection in the chicken. Avian Pathol 26(4):677–706
Hamzić E et al (2016) RNA sequencing-based analysis of the spleen transcriptome following infectious bronchitis virus infection of chickens selected for different mannose-binding lectin serum concentrations. BMC Genomics 17(1):1–13
Wang X et al (2006) Transcriptome of local innate and adaptive immunity during early phase of infectious bronchitis viral infection. Viral Immunol 19(4):768–774
Jordan B (2017) Vaccination against infectious bronchitis virus: a continuous challenge. Vet Microbiol 206:137–143
Meir R et al (2012) Immune responses to mucosal vaccination by the recombinant S1 and N proteins of infectious bronchitis virus. Viral Immunol 25(1):55–62
Xu P et al (2019) A Multi-omics study of chicken infected by nephropathogenic infectious bronchitis virus. Viruses 11(11):1070
Cong F et al (2013) Transcriptome analysis of chicken kidney tissues following coronavirus avian infectious bronchitis virus infection. BMC Genomics 14(1):1–13
Lin X et al (2015) Insights into human astrocyte response to H5N1 infection by microarray analysis. Viruses 7(5):2618–2640
Najafi H et al (2016) Pathogenicity characteristics of an Iranian variant-2 (IS-1494) like infectious bronchitis virus in experimentally infected SPF chickens. Acta Virol 60(4):393–399
Ren G et al (2020) Pathogenicity of a QX-like avian infectious bronchitis virus isolated in China. Poult Sci 99(1):111–118
Rauw W et al (1998) Undesirable side effects of selection for high production efficiency in farm animals: a review. Livest Prod Sci 56(1):15–33
Najafi H et al (2016) Molecular characterization of infectious bronchitis viruses isolated from broiler chicken farms in Iran, 2014–2015. Adv Virol 161(1):53–62
Reed LJ, Muench H (1938) A simple method of estimating fifty per cent endpoints. Am J Epidemiol 27(3):493–497
Dobin A et al (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29(1):15–21
Huang DW, Sherman BT, Lempicki RA (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37(1):1–13
Ye J et al (2012) Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13(1):1–11
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time qPCR and the 2− ΔΔCT method. Methods 25(4):402–408
Abdollahi H et al (2021) Coronavirus: proteomics analysis of chicken kidney tissue infected with variant 2 (IS-1494)-like avian infectious bronchitis virus. Adv Virol 166(1):101–113
Kanehisa M et al (2019) New approach for understanding genome variations in KEGG. Nucleic Acids Res 47(D1):D590–D595
Chhabra R, Chantrey J, Ganapathy K (2015) Immune responses to virulent and vaccine strains of infectious bronchitis viruses in chickens. Viral Immunol 28(9):478–488
Paul MS et al (2013) Immunostimulatory properties of Toll-like receptor ligands in chickens. Vet Immunol Immunopathol 152(3–4):191–199
Bowie AG, Haga IR (2005) The role of Toll-like receptors in the host response to viruses. Mol Immunol 42(8):859–867
Okabayashi T et al (2006) Cytokine regulation in SARS coronavirus infection compared to other respiratory virus infections. J Med Virol 78(4):417–424
Le Goffic R et al (2007) Cutting Edge: Influenza A virus activates TLR3-dependent inflammatory and RIG-I-dependent antiviral responses in human lung epithelial cells. J Immunol 178(6):3368–3372
Liu P et al (2007) Retinoic acid-inducible gene I mediates early antiviral response and Toll-like receptor 3 expression in respiratory syncytial virus-infected airway epithelial cells. J Virol 81(3):1401–1411
Guo X et al (2008) Molecular mechanisms of primary and secondary mucosal immunity using avian infectious bronchitis virus as a model system. Vet Immunol Immunopathol 121(3–4):332–343
Kameka AM et al (2014) Induction of innate immune response following infectious bronchitis corona virus infection in the respiratory tract of chickens. Virology 450:114–121
Chen S, Cheng A, Wang M (2013) Innate sensing of viruses by pattern recognition receptors in birds. Vet Res 44(1):1–12
Matsumoto M, Seya T (2008) TLR3: interferon induction by double-stranded RNA including poly (I: C). Adv Drug Deliv Rev 60(7):805–812
Fulton R et al (1997) Effect of Cytoxan®-induced heteropenia on the response of specific-pathogen-free chickens to infectious bronchitis. Avian Dis 41:511–518
Dar A et al (2005) Transcriptional analysis of avian embryonic tissues following infection with avian infectious bronchitis virus. Virus Res 110(1–2):41–55
Matthijs MG et al (2009) Course of infection and immune responses in the respiratory tract of IBV infected broilers after superinfection with E. coli. Vet Immunol Immunopathol 127(1–2):77–84
Garceau V et al (2010) Pivotal Advance: Avian colony-stimulating factor 1 (CSF-1), interleukin-34 (IL-34), and CSF-1 receptor genes and gene products. J Leukoc Biol 87(5):753–764
Dunkelberger JR, Song W-C (2010) Complement and its role in innate and adaptive immune responses. Cell Res 20(1):34–50
Liniger M et al (2012) Chicken cells sense influenza A virus infection through MDA5 and CARDIF signaling involving LGP2. J Virol 86(2):705–717
Loo Y-M, Gale M Jr (2011) Immune signaling by RIG-I-like receptors. Immunity 34(5):680–692
Keestra AM et al (2013) Unique features of chicken Toll-like receptors. Dev Comp Immunol 41(3):316–323
Barber MR et al (2010) Association of RIG-I with innate immunity of ducks to influenza. Proc Natl Acad Sci 107(13):5913–5918
Yang X et al (2018) Induction of innate immune response following introduction of infectious bronchitis virus (IBV) in the trachea and renal tissues of chickens. Microb Pathog 116:54–61
Rajsbaum R et al (2012) Species-specific inhibition of RIG-I ubiquitination and IFN induction by the influenza A virus NS1 protein. PLoS Pathog 8(11):e1003059
Acknowledgements
The authors gratefully acknowledge the Sheikh Bahaei National High-Performance Computing Center (SBNHPCC) for providing computing facilities and time. SBNHPCC is supported by the Presidential Office’s Scientific and Technological Department and Isfahan University of Technology.
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling Editor: Diego G. Diel.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ghobadian Diali, H., Hosseini, H., Fallah Mehrabadi, M.H. et al. Evaluation of viral load and transcriptome changes in tracheal tissue of two hybrids of commercial broiler chickens infected with avian infectious bronchitis virus: a comparative study. Arch Virol 167, 377–391 (2022). https://doi.org/10.1007/s00705-021-05322-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-021-05322-5