Abstract
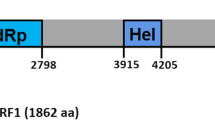
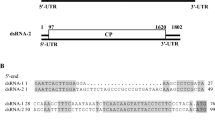
In this report, the full genome sequence of a novel mycovirus, designated as "Rhizoctonia solani partitivirus SM03" (RsPV-SM03), was determined in Rhizoctonia solani AG-4 HG III isolate SM03. RsPV-SM03 genome consists of two dsRNAs (dsRNA-1 and dsRNA-2), each of them contains one single open reading frame (ORF). ORF1 of dsRNA-1 encodes a putative RNA-dependent RNA polymerase (RdRp), while ORF2 of dsRNA-2 encodes a putative viral coat protein (CP). Phylogenetic analysis indicated that the RdRp and CP of RsPV-SM03 are closely related to those of other members of the genus Alphapartitivirus, family Partitiviridae, suggesting that RsPV-SM03 represents a novel species in the genus Alphapartitivirus.



Similar content being viewed by others
References
Marquez LM, Redman RS, Rodriguez RJ, Roossinck MJ (2007) A virus in a fungus in a plant: three-way symbiosis required for thermal tolerance. Science 315:513–515
Lyu R, Zhang Y, Tang Q, Li Y, Cheng J, Fu Y, Chen T, Jiang D, Xie J (2018) Two alphapartitiviruses co-infecting a single isolate of the plant pathogenic fungus Rhizoctonia solani. Arch Virol 163:515–520
Lin YH, Fujita M, Chiba S, Hyodo K, Andika IB, Suzuki N, Kondo H (2019) Two novel fungal negative-strand RNA viruses related to mymonaviruses and phenuiviruses in the shiitake mushroom (Lentinula edodes). Virology 533:125–136
Xie J, Jiang D (2014) New insights into mycoviruses and exploration for the biological control of crop fungal diseases. Annu Rev Phytopathol 52:45–68
Zhang T, Li N, Yuan Y, Cao Q, Chen Y, Tan B, Li G, Liu D (2019) Blue-white colony selection of virus-infected isogenic recipients based on a chrysovirus isolated from penicillium italicum. Virol Sin 34:688–700
Stalpes JA, Andersen TF (1996) A synopsis of the taxonomy of teleomorphs connected with Rhizoctonia S.L. In: Sneh B, Jabaji-Hare S, Neate S, Dijst G (eds) Rhizoctonia species taxonomy, molecular biology, ecology, pathology, and disease control. pp 50–63
Carling DE, Baird RE, Gitaitis RD, Brainard KA, Kuninaga S (2002) Characterization of AG-13, a newly reported anastomosis group of Rhizoctonia solani. Phytopathology 92:893–899
Sharon M, Sneh B, Kuninaga S, Hyakumachi M (2006) The advancing identification and classification of Rhizoctonia spp. using molecular and biotechnological methods compared with the classical anastomosis grouping. Mycoscience 47:299–316
DE Carling KS, Brainard KA (2002) Hyphal anastomosis reactions, rDNA-internal transcribed spacer sequences, and virulence levels among subsets of Rhizoctonia solani anastomosis group-2 (AG-2) and AG-BI. Phytopathology 92:43–50
Bharathan N, Saso H, Gudipati L, Bharathan S, Whited K (2005) Double-stranded RNA: distribution and analysis among isolates of Rhizoctonia solani AG-2 to -13. Plant Pathol 54:196–203
Zanzinger DH, Bandy BP, Tavantzis SM (1984) High frequency of finding double-stranded RNA in naturally occurring isolates of Rhizoctonia solani. J Gen Virol 65:1601–1605
Li Y (2015) The establishment of a new anastomosis group AG-V and research of RNA viruses from Rhizoctonia spp., Dissertation, Yunnan Agricultural University
Abdoulaye AH, Foda MF, Kotta-Loizou I (2019) Viruses infecting the plant pathogenic fungus Rhizoctonia solani. Viruses. https://doi.org/10.3390/v11121113
Marzano SL, Nelson BD, Ajayi-Oyetunde O, Bradley CA, Hughes TJ, Hartman GL, Eastburn DM, Domier LL (2016) Identification of diverse mycoviruses through metatranscriptomics characterization of the viromes of five major fungal plant pathogens. J Virol 90:6846–6863
Zheng L, Zhang M, Chen Q, Zhu M, Zhou E (2014) A novel mycovirus closely related to viruses in the genus Alphapartitivirus confers hypovirulence in the phytopathogenic fungus Rhizoctonia solani. Virology 456–457:220–226
Liu C, Zeng M, Zhang M, Shu C, Zhou E (2018) Complete nucleotide sequence of a partitivirus from Rhizoctonia solani AG-1 IA strain C24. Viruses. https://doi.org/10.3390/v10120703
Zhang M, Zheng L, Liu C, Shu C, Zhou E (2018) Characterization of a novel dsRNA mycovirus isolated from strain A105 of Rhizoctonia solani AG-1 IA. Arch Virol 163:427–430
Strauss EE, Lakshman DK, Tavantzis SM (2000) Molecular characterization of the genome of a partitivirus from the basidiomycete Rhizoctonia solani. J Gen Virol 81:549–555
Chen Y, Gai X, Chen R, Li C, Zhao G, Xia Z, Zou C, Zhong J (2019) Characterization of three novel betapartitiviruses co-infecting the phytopathogenic fungus Rhizoctonia solani. Virus Res 270:197649
Morris T, Dodds JA (1979) Isolation and analysis of double-stranded RNA from virus-infected plant and fungal tissue. Phytopathology 69:854–858
Darissa O, Willingmann P, Adam G (2010) Optimized approaches for the sequence determination of double-stranded RNA templates. J Virol Methods 169:397–403
Froussard P (1992) A random-PCR method (rPCR) to construct whole cDNA library from low amounts of RNA. Nucleic Acids Res 20:11
Potgieter AC, Page NA, Liebenberg J, Wright IM, Landt O, van Dijk AA (2009) Improved strategies for sequence-independent amplification and sequencing of viral double-stranded RNA genomes. J Gen Virol 90:1423–1432
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Acknowledgements
This research was supported by Kunming University (YJL16006), Department of Science and Technology of Yunnan Province (2018FH001-032, 2018FH001-052, 2018FD080), and the National Natural Science Foundation of China (31960525).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to declare that are relevant to the content of this article.
Ethical approval
The authors have no relevant financial or non-financial interests to disclose. All authors certify that they have no affiliations with or involvement in any organization or entity with any financial interest or non-financial interest in the subject matter or materials discussed in this article. The authors have no financial or proprietary interests in any material discussed in this article.
Additional information
Handling Editor: Massimo Turina.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Li, Y., Chen, W., Niu, Y. et al. Complete nucleotide sequence of a novel alphapartitivirus from Rhizoctonia solani AG-4 HG III isolate SM03. Arch Virol 167, 953–957 (2022). https://doi.org/10.1007/s00705-021-05261-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-021-05261-1