Abstract
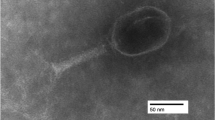
In this study, a novel Escherichia coli-specific bacteriophage, vB_EcoM_IME392, was isolated from chicken farm sewage in Qingdao, China. The genome of IME392 was found by next-generation sequencing to be 116,460 base pairs in length with a G+C content of 45.4% (GenBank accession number MH719082). BLASTn results revealed that only 2% of the genome sequence of IME392 shows sequence similarity to known phage sequences in the GenBank database, which indicates that IME392 is a novel bacteriophage. Transmission electron microscopy showed that IME392 belongs to the family Myoviridae. The host range, the multiplicity of infection, and a one-step growth curve were also determined.





Similar content being viewed by others
References
Andres D, Baxa U, Hanke C, Seckler R, Barbirz S (2010) Carbohydrate binding of Salmonella phage P22 tailspike protein and its role during host cell infection. Biochem Soc Trans 38:1386–1389
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O (2008) The RAST Server: rapid annotations using subsystems technology. BMC Genom 9:75
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120
Chen X, Zhou L, Tian K, Kumar A, Singh S, Prior BA, Wang Z (2013) Metabolic engineering of Escherichia coli: a sustainable industrial platform for bio-based chemical production. Biotechnol Adv 31:1200–1223
Chibani-Chennoufi S, Canchaya C, Bruttin A, Brüssow H (2004) Comparative genomics of the T4-Like Escherichia coli phage JS98: implications for the evolution of T4 phages. J Bacteriol 186:8276–8286
Cohen SN, Chang AC, Boyer HW, Helling RB (1973) Construction of biologically functional bacterial plasmids in vitro. Proc Natl Acad Sci USA 70:3240–3244
Crick FH, Barnett L, Brenner S, Watts-Tobin RJ (1961) General nature of the genetic code for proteins. Nature 192:1227–1232
Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA (2005) Diversity of the human intestinal microbial flora. Science 308:1635–1638
Fiers W, Contreras R, Duerinck F, Haegeman G, Iserentant D, Merregaert J, Min Jou W, Molemans F, Raeymaekers A, Van den Berghe A, Volckaert G, Ysebaert M (1976) Complete nucleotide sequence of bacteriophage MS2 RNA: primary and secondary structure of the replicase gene. Nature 260:500–507
Fu P, Zhao Q, Shi L, Xiong Q, Ren Z, Xu H, Chai S, Xu Q, Sun X, Sang M (2021) Identification and characterization of two bacteriophages with lytic activity against multidrug-resistant Escherichia coli. Virus Res 291:198196
Guo Z, Huang J, Yan G, Lei L, Wang S, Yu L, Zhou L, Gao A, Feng X, Han W, Gu J, Yang J (2017) Identification and characterization of Dpo42, a novel depolymerase derived from the Escherichia coli phage vB_EcoM_ECOO78. Front Microbiol 8:1460
Huang CJ, Lin H, Yang X (2012) Industrial production of recombinant therapeutics in Escherichia coli and its recent advancements. J Ind Microbiol Biotechnol 39:383–399
Jassim SA, Limoges RG (2014) Natural solution to antibiotic resistance: bacteriophages “The Living Drugs.” World J Microbiol Biotechnol 30:2153–2170
Jaureguy F, Landraud L, Passet V, Diancourt L, Frapy E, Guigon G, Carbonnelle E, Lortholary O, Clermont O, Denamur E, Picard B, Nassif X, Brisse S (2008) Phylogenetic and genomic diversity of human bacteremic Escherichia coli strains. BMC Genom 9:560
Kaper JB, Nataro JP, Mobley HL (2004) Pathogenic Escherichia coli. Nat Rev Microbiol 2:123–140
Kauffman KM, Brown JM, Sharma RS, VanInsberghe D, Elsherbini J, Polz M, Kelly L (2018) Viruses of the Nahant Collection, characterization of 251 marine Vibrionaceae viruses. Sci Data 5:180114
Kim B, Park H, Na D, Lee SY (2014) Metabolic engineering of Escherichia coli for the production of phenol from glucose. Biotechnol J 9:621–629
Kutter EM, Skutt-Kakaria K, Blasdel B, El-Shibiny A, Castano A, Bryan D, Kropinski AM, Villegas A, Ackermann HW, Toribio AL, Pickard D, Anany H, Callaway T, Brabban AD (2011) Characterization of a ViI-like phage specific to Escherichia coli O157:H7. Virol J 8:430
Laxminarayan R, Amábile-Cuevas CF, Cars O, Evans T, Heymann DL, Hoffman S, Holmes A, Mendelson M, Sridhar D, Woolhouse M, Røttingen JA (2016) UN High-Level Meeting on antimicrobials–what do we need? Lancet 388:218–220
Linn S, Arber W (1968) Host specificity of DNA produced by Escherichia coli, X. In vitro restriction of phage fd replicative form. Proc Natl Acad Sci USA 59:1300–1306
Liu T, Khosla C (2010) Genetic engineering of Escherichia coli for biofuel production. Annu Rev Genet 44:53–69
Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu LF, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu JH, Shen J (2016) Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168
Luria SE, Delbrück M (1943) Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28:491–511
O'Neill J (2016) Tackling drug-resistant infections globally: final report and recommendations. Government of the United Kingdom
Opella SJ, Stewart PL, Valentine KG (1987) Protein structure by solid-state NMR spectroscopy. Q Rev Biophys 19:7–49
Petrov VM, Nolan JM, Bertrand C, Levy D, Desplats C, Krisch HM, Karam JD (2006) Plasticity of the gene functions for DNA replication in the T4-like phages. J Mol Biol 361:46–68
Pitout JD (2012) Extraintestinal pathogenic Escherichia coli: an update on antimicrobial resistance, laboratory diagnosis and treatment. Expert Rev Anti Infect Ther 10:1165–1176
Russo TA, Johnson JR (2003) Medical and economic impact of extraintestinal infections due to Escherichia coli: focus on an increasingly important endemic problem. Microbes Infect 5:449–456
Sanger F, Air GM, Barrell BG, Brown NL, Coulson AR, Fiddes CA, Hutchison CA, Slocombe PM, Smith M (1977) Nucleotide sequence of bacteriophage phi X174 DNA. Nature 265:687–695
Wick RR, Schultz MB, Zobel J, Holt KE (2015) Bandage: interactive visualization of de novo genome assemblies. Bioinformatics 31:3350–3352
Williams GJ, Thorson JS (2009) Natural product glycosyltransferases: properties and applications. Adv Enzymol Relat Areas Mol Biol 76:55–119
Yuan W, Zhang Y, Wang G, Bai J, Wang X, Li Y, Jiang P (2016) Genomic and proteomic characterization of SE-I, a temperate bacteriophage infecting Erysipelothrix rhusiopathiae. Arch Virol 161:3137–3150
Zhang Q, Xing S, Sun Q, Pei G, Cheng S, Liu Y, An X, Zhang X, Qu Y, Tong Y (2017) Characterization and complete genome sequence analysis of a novel virulent Siphoviridae phage against Staphylococcus aureus isolated from bovine mastitis in Xinjiang, China. Virus Genes 53:464–476
Zhou Y, Bao H, Zhang H, Wang R (2015) Isolation and Characterization of Lytic Phage vB_EcoM_JS09 against Clinically Isolated Antibiotic-Resistant Avian Pathogenic Escherichia coli and Enterotoxigenic Escherichia coli. Intervirology 58:218–231
Acknowledgements
This research was supported by the Key Project of Beijing University of Chemical Technology (XK1803-06), the National Key Research and Development Program of China (2018YFA0903000, 2020YFC2005405, 2020YFA0712100, 2020YFC0840805), Funds for First-Class Discipline Construction (XK1805), the Inner Mongolia Key Research and Development Program (2019ZD006), the National Natural Science Foundation of China (81672001, 81621005), the NSFC-MFST Project (China-Mongolia) (31961143024), and Fundamental Research Funds for Central Universities (BUCTRC201917, BUCTZY2022).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Handling Editor: Johannes Wittmann.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yunjia Hu, Shanwei Tong and Ping Li are contributed equally to this work.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hu, Y., Tong, S., Li, P. et al. Characterization and genome sequence of the genetically unique Escherichia bacteriophage vB_EcoM_IME392. Arch Virol 166, 2505–2520 (2021). https://doi.org/10.1007/s00705-021-05160-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-021-05160-5