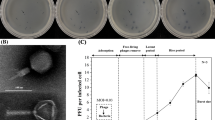
Abstract
Vibrio parahaemolyticus is a major foodborne pathogen and is also pathogenic to shrimp. Due to the emergence of multidrug-resistant V. parahaemolyticus strains, bacteriophages have shown promise as antimicrobial agents that could be used for controlling antibiotic-resistant strains. Here, a V. parahaemolyticus phage, vB_VpaP_MGD2, was isolated from a clam (Meretrix meretrix) and further characterized to evaluate its potential capability for biocontrol. Podophage vB_VpaP_MGD2 had a wide host range and was able to lyse 27 antibiotic-resistant V. parahaemolyticus strains. A one-step growth curve showed that vB_VpaP_MGD2 has a short latent period of 10 min and a large burst size of 244 phages per cell. Phage vB_VpaP_MGD2 was able to tolerate a wide range of temperature (30 °C-50 °C) and pH (pH 3-pH 10). Two multidrug-resistant strains (SH06 and SA411) were suppressed by treatment with phage vB_VpaP_MGD2 at a multiplicity of infection of 100 for 24 h without apparent regrowth of bacterial populations. The frequency of mutations causing bacteriophage resistance was relatively low (3.1 × 10-6). Phage vB_VpaP_MGD2 has a double-stranded DNA with a genome size of 45,105 bp. Among the 48 open reading frames annotated in the genome, no lysogenic genes or virulence genes were detected. Sequence comparisons suggested that vB_VpaP_MGD2 is a member of a new species in the genus Zindervirus within the subfamily Autographivirinae. This is the first report of a member of the genus Zindervirus that can infect V. parahaemolyticus. These findings suggest that vB_VpaP_MGD2 may be a candidate biocontrol agent against early mortality syndrome/acute hepatopancreatic necrosis disease (EMS/AHPND) caused by multidrug-resistant V. parahaemolyticus in shrimp production.






Similar content being viewed by others
References
Han N, Mizan MFR, Jahid IK, Ha SD (2016) Biofilm formation by Vibrio parahaemolyticus on food and food contact surfaces increases with rise in temperature. Food Control 70:161–166. https://doi.org/10.1016/j.foodcont.2016.05.054
Centers for Disease Control and Prevention (2019) Vibrio illness (Vibriosis): Vibrio parahaemolyticus. http://www.cdc.gov/vibrio/vibriop.html. Accessed 4 Aug 2019
Han X, Zhang H, Cao M, Shen X (2015) Analysis on current pollution of Vibrio parahemolyticus in seafood in China and its controlling strategy. Food Ferment Ind 7:263–267. https://doi.org/10.13995/j.cnki.11-1802/ts.201507048
Kongrueng J, Srinitiwarawong K, Nishibuchi M, Mittraparp-Arthorn P, Vuddhakul V (2018) Characterization and CRISPR-based genotyping of clinical trh-positive Vibrio parahaemolyticus. Gut Pathog 10:48. https://doi.org/10.1186/s13099-018-0275-4
Yang Q, Dong X, Xie G, Fu S, Zou P, Sun J, Wang Y, Huang J (2019) Comparative genomic analysis unravels the transmission pattern and intra-species divergence of acute hepatopancreatic necrosis disease (AHPND)-causing Vibrio parahaemolyticus strains. Mol Genet Genomics 294:1007–1022. https://doi.org/10.1007/s00438-019-01559-7
Shaw KS, Goldstein RER, He X, Jacobs JM, Crump BC, Sapkota AR (2014) Antimicrobial susceptibility of Vibrio vulnificus and Vibrio parahaemolyticus recovered from recreational and commercial areas of Chesapeake Bay and Maryland Coastal Bays. PLoS ONE 9:e89616. https://doi.org/10.1371/journal.pone.0089616
Xu X, Cheng J, Wu Q, Zhang J, Xie T (2016) Prevalence, characterization, and antibiotic susceptibility of Vibrio parahaemolyticus isolated from retail aquatic products in North China. BMC Microbiol 16:32. https://doi.org/10.1186/s12866-016-0650-6
Obaidat MM, Salman AEB, Roess AA (2017) Virulence and antibiotic resistance of Vibrio parahaemolyticus isolates from seafood from three developing countries and of worldwide environmental, seafood, and clinical isolates from 2000 to 2017. J Food Prot 80:2060–2067. https://doi.org/10.4315/0362-028x.jfp-17-156
Lee Y, Choi Y, Lee S, Lee H, Kim S, Ha J, Lee J, Oh H, Kim Y, Yoon Y (2019) Occurrence of pathogenic Vibrio parahaemolyticus in seafood distribution channels and their antibiotic resistance profiles in S. Korea. Lett Appl Microbiol 68:128–133. https://doi.org/10.1111/lam.13099
Letchumanan V, Yin WF, Lee LH, Chan KG (2015) Prevalence and antimicrobial susceptibility of Vibrio parahaemolyticus isolated from retail shrimps in Malaysia. Front Microbiol 6:33. https://doi.org/10.3389/fmicb.2015.00033
Gräslund S, Bengtsson BE (2001) Chemicals and biological products used in south-east Asian shrimp farming, and their potential impact on the environment—a review. Sci Total Environ 280:93–131. https://doi.org/10.1016/s0048-9697(01)00818-x
Adams A (2019) Progress, challenges and opportunities in fish vaccine development. Fish Shellfish Immunol 90:210–214. https://doi.org/10.1016/j.fsi.2019.04.066
Wittebole X, De Roock S, Opal SM (2014) A historical overview of bacteriophage therapy as an alternative to antibiotics for the treatment of bacterial pathogens. Virulence 5:226–235. https://doi.org/10.4161/viru.25991
Rao BM, Lalitha KV (2015) Bacteriophage for aquaculture: are they beneficial or inimical. Aquaculture 437:146–154. https://doi.org/10.1016/j.aquaculture.2014.11.039
Kazi M, Annapure US (2016) Bacteriophage biocontrol of foodborne pathogens. J Food Sci Technol 53:1355–1362. https://doi.org/10.1007/s13197-015-1996-8
Martinez-Diaz SF, Hipolito-Morales A (2013) Efficacy of phage therapy to prevent mortality during the vibriosis of brine shrimp. Aquaculture 400–401:120–124. https://doi.org/10.1016/j.aquaculture.2013.03.007
Wang YH, Barton M, Elliott L, Li XX, Abraham S, O’Dea M, Munro J (2017) Bacteriophage therapy for the control of Vibrio harveyi in greenlip abalone (Haliotis laevigata). Aquaculture 473:251–258. https://doi.org/10.1016/j.aquaculture.2017.01.003
Khairnar K, Raut MP, Chandekar RH, Sanmukh SG, Paunikar WN (2013) Novel bacteriophage therapy for controlling metallo-beta-lactamase producing Pseudomonas aeruginosa infection in catfish. BMC Vet Res 9:264. https://doi.org/10.1186/1746-6148-9-264
Nikapitiya C, Chandrarathna HPSU, Dananjaya SHS, De Zoysa M, Lee J (2020) Isolation and characterization of phage (ETP-1) specific to multidrug resistant pathogenic Edwardsiella tarda and its in vivo biocontrol efficacy in zebrafish (Danio rerio). Biologicals 63:14–23. https://doi.org/10.1016/j.biologicals.2019.12.006
Le TS, Nguyen TH, Vo HP, Doan VC, Nguyen HL, Tran MT, Tran TT, Southgate PC, Kurtböke Dİ (2018) Protective effects of bacteriophages against Aeromonas hydrophila species causing motile Aeromonas Septicemia (MAS) in striped catfish. Antibiotics (Basel) 7:16. https://doi.org/10.3390/antibiotics7010016
Matsuoka S, Hashizume T, Kanzaki H, Iwamoto E, Park SC, Yoshida T, Nakai T (2007) Phage therapy against beta-hemolytic streptococcicosis of Japanese flounder Paralichthys olivaceus. Fish Pathol 42:181–189. https://doi.org/10.3147/jsfp.42.181
García-Anaya MC, Sepulveda DR, Sáenz-Mendoza AI, Rios-Velasco C, Zamudio-Flores PB, Acosta-Muñiz CH (2020) Phages as biocontrol agents in dairy products. Trends Food Sci Tech 95:10–20. https://doi.org/10.1016/j.tifs.2019.10.006
Lomelí-Ortega CO, Martínez-Díaz SF (2014) Phage therapy against Vibrio parahaemolyticus infection in the whiteleg shrimp (Litopenaeus vannamei) larvae. Aquaculture 434:208–211. https://doi.org/10.1016/j.aquaculture.2014.08.018
Rong R, Lin H, Wang J, Khan MN, Li M (2014) Reductions of Vibrio parahaemolyticus in oysters after bacteriophage application during depuration. Aquaculture 418:171–176. https://doi.org/10.1016/j.aquaculture.2013.09.028
Zhang H, Yang Z, Zhou Y, Bao H, Wang R, Li T, Pang M, Sun L, Zhou X (2018) Application of a phage in decontaminating Vibrio parahaemolyticus in oysters. Int J Food Microbiol 275:24–31. https://doi.org/10.1016/j.ijfoodmicro.2018.03.027
Yin Y, Ni P, Liu D, Yang S, Almeida A, Guo Q, Zhang Z, Deng L, Wang D (2019) Bacteriophage potential against Vibrio parahaemolyticus biofilms. Food Control 98:156–163. https://doi.org/10.1016/j.foodcont.2018.11.034
Jun JW, Shin TH, Kim JH, Shin SP, Han JE, Heo GJ, De Zoysa M, Shin GW, Chai JY, Park SC (2014) Bacteriophage therapy of a Vibrio parahaemolyticus infection caused by a multiple-antibiotic-resistant O3:K6 pandemic clinical strain. J Infect Dis 210:72–78. https://doi.org/10.1093/infdis/jiu059
Lee LH, Ab Mutalib NS, Law JW, Wong SH, Letchumanan V (2018) Discovery on antibiotic resistance patterns of Vibrio parahaemolyticus in Selangor reveals carbapenemase producing Vibrio parahaemolyticus in marine and freshwater fish. Front Microbiol 9:2513. https://doi.org/10.3389/fmicb.2018.02513
Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL (2012) Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18:268–281. https://doi.org/10.1111/j.1469-0691.2011.03570.x
Topka G, Bloch S, Nejman-Faleńczyk B, Gąsior T, Jurczak-Kurek A, Necel A, Dydecka A, Richert M, Węgrzyn G, Węgrzyn A (2019) Characterization of bacteriophage vB-EcoS-95, isolated from urban sewage and revealing extremely rapid lytic development. Front Microbiol 9:3326. https://doi.org/10.3389/fmicb.2018.03326
Endersena L, Buttimera C, Nevina E, Coffeya A, Neveb H, Oliveira H, Lavigne R, O’Mahony J (2017) Investigating the biocontrol and anti-biofilm potential of a three phage cocktail against Cronobacter sakazakii in different brands of infant formula. Int J Food Microbiol 275:24–31. https://doi.org/10.1016/j.ijfoodmicro.2017.04.009
Chen L, Yuan S, Liu Q, Mai G, Yang J, Deng D, Zhang B, Liu C, Ma Y (2018) In vitro design and evaluation of phage cocktails against Aeromonas salmonicida. Front Microbiol 9:1476. https://doi.org/10.3389/fmicb.2018.01476
Sasikala D, Srinivasan P (2016) Characterization of potential lytic bacteriophage against Vibrio alginolyticus and its therapeutic implications on biofilm dispersal. Microb Pathogenesis 101:24–35. https://doi.org/10.1016/j.micpath.2016.10.017
Ciacci N, D’Andrea MM, Marmo P, Demattè E, Amisano F, Di Pilato V, Fraziano M, Lupetti P, Rossolini GM, Thaller MC (2018) Characterization of vB_Kpn_F48, a newly discovered lytic bacteriophage for Klebsiella pneumoniae of sequence type 101. Viruses 10:482. https://doi.org/10.3390/v10090482
Alves DR, Gaudion A, Bean JE, Perez Esteban P, Arnot TC, Harper DR, Kot W, Hansen LH, Enright MC, Jenkins AT (2014) Combined use of bacteriophage K and a novel bacteriophage to reduce Staphylococcus aureus biofilm formation. Appl Environ Microb 80:6694–6703. https://doi.org/10.1128/AEM.01789-14
Taha OA, Connerton PL, Connerton IF, El-Shibiny A (2018) Bacteriophage ZCKP1: a potential treatment for Klebsiella pneumoniae isolated from diabetic foot patients. Front Microbiol 9:2127. https://doi.org/10.3389/fmicb.2018.02127
Coil D, Jospin G, Darling AE (2015) A5-MiSeq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics 31:587–589. https://doi.org/10.1093/bioinformatics/btu661
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. https://doi.org/10.1089/cmb.2012.0021
Garneau JR, Depardieu F, Fortier LC, Bikard D, Monot M (2017) PhageTerm: a tool for fast and accurate determination of phage termini and packaging mechanism using next-generation sequencing data. Sci Rep 7:8292. https://doi.org/10.1038/s41598-017-07910-5
Schattner P, Brooks AN, Lowe TM (2005) The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res 33:W686–W689. https://doi.org/10.1093/nar/gki366
Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV (2012) Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. https://doi.org/10.1093/jac/dks261
Chen L, Zheng D, Liu B, Yang J, Jin Q (2016) VFDB 2016: hierarchical and refined dataset for big data analysis—10 years on. Nucleic Acids Res 44:D694–D697. https://doi.org/10.1093/nar/gkv1239
Stothard P, Wishart DS (2005) Circular genome visualization and exploration using CGView. Bioinformatics 21:537–539. https://doi.org/10.1093/bioinformatics/bti054
Ferri M, Ranucci E, Romagnoli P, Giaccone V (2017) Antimicrobial resistance: a global emerging threat to public health systems. Crit Rev Food Sci Nutr 57:2857–2876. https://doi.org/10.1080/10408398.2015.1077192
Letchumanan V, Chan KG, Pusparajah P, Saokaew S, Duangjai A, Goh BH, Ab Mutalib NS, Lee LH (2016) Insights into bacteriophage application in controlling Vibrio species. Front Microbiol 7:1114. https://doi.org/10.3389/fmicb.2016.01114
Barros J, Melo LDR, Poeta P, Igrejas G, Ferraz MP, Azeredo J, Monteiro FJ (2019) Lytic bacteriophages against multidrug-resistant Staphylococcus aureus, Enterococcus faecalis and Escherichia coli isolates from orthopaedic implant-associated infections. Int J Antimicrob Agents 54:329–337. https://doi.org/10.1016/j.ijantimicag.2019.06.007
Łubowska N, Grygorcewicz B, Kosznik-Kwaśnicka K, Zauszkiewicz-Pawlak A, Węgrzyn A, Dołęgowska B, Piechowicz L (2019) Characterization of the three new kayviruses and their lytic activity against multidrug-resistant Staphylococcus aureus. Microorganisms 7:471. https://doi.org/10.3390/microorganisms7100471
Gupta R, Prasad Y (2011) Efficacy of polyvalent bacteriophage P-27/HP to control multidrug resistant Staphylococcus aureus associated with human infections. Curr Microbiol 62:255. https://doi.org/10.1007/s00284-010-9699-x
Amarillas L, Rubí-Rangel L, Chaidez C, González-Robles A, Lightbourn-Rojas L, León-Félix J (2017) Isolation and characterization of phiLLS, a novel phage with potential biocontrol agent against multidrug-resistant Escherichia coli. Front Microbiol 8:1355. https://doi.org/10.3389/fmicb.2017.01355
Lal TM, Sano M, Ransangan J (2017) Isolation and characterization of large marine bacteriophage (Myoviridae), VhKM4 infecting Vibrio harveyi. J Aquat Anim Health 29:26–30. https://doi.org/10.1080/08997659
Wong HC, Wang TY, Yang CW, Tang CT, Ying CW, Wang CH, Chang WH (2019) Characterization of a lytic vibriophage VP06 of Vibrio parahaemolyticus. Res Microbiol 170:13–23. https://doi.org/10.1016/j.resmic.2018.07.003
Lal TM, Sano M, Ransangan J (2016) Genome characterization of a novel vibriophage VpKK5 (Siphoviridae) specific to fish pathogenic strain of Vibrio parahaemolyticus. J Basic Microbiol 56:872–888. https://doi.org/10.1002/jobm.201500611
Stalin N, Srinivasan P (2016) Characterization of Vibrio parahaemolyticus and its specific phage from shrimp pond in Palk Strait, South East coast of India. Biologicals 44:526–533. https://doi.org/10.1016/j.biologics.2016.08.003
Adriaenssens E, Brister JR (2017) How to name and classify your phage: an informal guide. Viruses 9:70. https://doi.org/10.3390/v9040070
Zhao F, Sun H, Zhou X, Liu G, Li M, Wang C, Liu S, Zhuang Y, Tong Y, Ren H (2019) Characterization and genome analysis of a novel bacteriophage vB_SpuP_Spp16 that infects Salmonella enterica serovar pullorum. Virus Genes 55:532–540. https://doi.org/10.1007/s11262-019-01664-0
Funding
This work was supported by the Shanghai Agriculture Applied Technology Development Program, China (grant no. 2019-02-08-00-10-F01149).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Yanzi Cao declares that she has no conflict of interest. Yujie Zhang declares that she has no conflict of interest. Weiqing Lan declares that he has no conflict of interest. Xiaohong Sun declares that she has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Handling Editor: Johannes Wittmann.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Cao, Y., Zhang, Y., Lan, W. et al. Characterization of vB_VpaP_MGD2, a newly isolated bacteriophage with biocontrol potential against multidrug-resistant Vibrio parahaemolyticus. Arch Virol 166, 413–426 (2021). https://doi.org/10.1007/s00705-020-04887-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-020-04887-x