Abstract
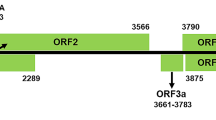
Analysis of a loquat tree with leaf curl symptoms by deep sequencing revealed a novel virus with a single-stranded RNA genome, for which the name “loquat virus A” (LoVA) was proposed. The complete genome sequence comprised 7553 nucleotides (nt) and an additional poly(A) tail at the 3′ terminus. Sequence comparisons of LoVA showed moderate similarity to cherry virus A (CVA), currant virus A (CuVA), and mume virus A (MuVA), which are members of the genus Capillovirus in the family Betaflexiviridae. Phylogenetic analysis of the full-genome nt sequence and replicase-like protein supported the placement of LoVA within the genus Capillovirus. However, it has a distinct genome organization, differing from recognized capilloviruses, as it contains three open reading frames (ORFs), with the coat protein (CP) expressed separately from the replication-associated protein (RP) rather than being encoded in the same ORF. This indicates that LoVA is a novel member of the genus Capillovirus in the family Betaflexiviridae with a distinct genomic organization.


Similar content being viewed by others
References
Roossinck MJ, Martin DP, Roumagnac P (2015) Plant virus metagenomics: advances in virus discovery. Phytopathology 105:716–727
Adams MJ, Candresse T, Hammond J, Kreuze JF, Martelli GP, Namba S, Pearson MN, Ryu KH, Saldarelli P, Yoshikawa N (2012) Family Betaflexiviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (eds) Virus taxonomy: ninth report of the international committee on taxonomy of viruses. Elsevier Academic Press, San Diego, pp 920–941
ICTV Master Species List (version MSL34). https://talk.ictvonline.org/taxonomy/vmr/m/vmr-file-repository/8287. Accessed Apr 2019
Tatineni S, Afunian MR, Gowda S, Hilf ME, Bar-Joseph M, Dawson WO (2009) Characterization of the 5′- and 3′-terminal subgenomic RNAs produced by a capillovirus: evidence for a CP subgenomic RNA. Virology 385:521–528
Sgroi F, Di Trapani AM, Squatrito R, Tudisca S, Testa R (2014) Economic assessment of Sicilian loquat orchards. Acta Hortic 1092:33–36. https://doi.org/10.17660/ActaHortic.2015.1092.3
Liu Q, Xuan Z, Wu J, Qiu Y, Li M, Zhang S, Wu D, Li R, Cao M (2019) Loquat is a new natural host of apple stem grooving virus and apple chlorotic leaf spot virus in China. Plant Dis. https://doi.org/10.1094/PDIS-04-19-0721-PDN
Velasco R, Zharkikh A, Affourtit J, Dhingra A, Cestaro A, Kalyanaraman A, Fontana P, Bhatnagar SK, Troggio M, Pruss D et al (2010) The genome of the domesticated apple (Malus × domestica Borkh.). Nat Genet 42:833–839
Cao M, Li P, Zhang S, Yang F, Zhou Y, Wang X, Li R, Li Z (2018) Molecular characterization of a novel citrivirus from citrus using next-generation sequencing. Arch Virol 163:3479–3482
Petrzik K, Pribylova J, Koloniuk I, Spak J (2016) Molecular characterization of a novel capillovirus from red currant. Arch Virol 161:1083–1086
Mushegian AR, Elena SF (2015) Evolution of plant virus movement proteins from the 30K superfamily and of their homologs integrated in plant genomes. Virology 476:304–315
Simon-Loriere E, Holmes EC (2011) Why do RNA viruses recombine? Nat Rev Microbiol 9:617–626
Martin DP, Murrell B, Golden M, Khoosal A, Muhire B (2015) RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol 1:vev003
Acknowledgements
This research was supported by the Intergovernmental International Science, Technology and Innovation (STI) Collaboration Key Project of China′s National Key R&D Program (NKP) (2017YFE0110900), Fundamental Research Funds for the Central Universities (XDJK2018AA002), Chongqing Research Program of Basic Research and Frontier Technology (cstc2017jcyjBX0016), Overseas Expertise Introduction Project for Discipline Innovation (111 Center) (B18044). We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript. We thank the editor and three anonymous reviewers for their constructive comments and suggestions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
I have read and abided by the statement of ethical standards for manuscripts submitted to Archives of Virology.
Conflict of interest
All authors declare they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animal performed by any of the authors.
Additional information
Handling Editor: Sead Sabanadzovic.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
Scheme of rapid amplification of cDNA ends (RACE), RT-PCR amplification of overlapped fragments, and Sanger sequencing (JPEG 75 kb)
Rights and permissions
About this article
Cite this article
Liu, Q., Yang, L., Xuan, Z. et al. Complete nucleotide sequence of loquat virus A, a member of the family Betaflexiviridae with a novel genome organization. Arch Virol 165, 223–226 (2020). https://doi.org/10.1007/s00705-019-04444-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-019-04444-1