Abstract
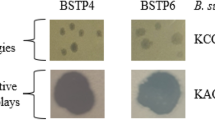
Bacillus subtilis-infecting phage BSP38 was isolated from a sewage sample. Morphologically, BSP38 was found to be similar to members of the subfamily Spounavirinae, family Myoviridae. Its genome is 153,268 bp long with 41.8% G+C content and 254 putative open reading frames (ORFs) as well as six tRNAs. A distinguishing feature for this phage among the reported B. subtilis-infecting phages is the presence of an encoding ORF, putative tRNAHis guanylyltransferase-like protein. Genomic comparisons with the other reported phages strongly suggest that BSP38 should be considered a member of a new genus in the subfamily Spounavirinae.


Similar content being viewed by others
References
Adriaenssens E, Brister JR (2017) How to name and classify your phage: an informal guide. Viruses 9:70
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M (2008) The RAST Server: rapid annotations using subsystems technology. BMC Genom 9:75
Barylski J, Nowicki G, Goździcka-Józefiak A (2014) The discovery of phiAGATE, a novel phage infecting Bacillus pumilus, leads to new insights into the phylogeny of the subfamily Spounavirinae. PLoS One 9:e86632
Besemer J, Lomsadze A, Borodovsky M (2001) GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res 29:2607–2618
Castresana J (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17:540–552
Chevenet F, Brun C, Bañuls A-L, Jacq B, Christen R (2006) TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinform 7:439
Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36:W465–W469
Edgar RC (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinform 5:113
Ghosh K, Kang HS, Hyun WB, Kim KP (2018) High prevalence of Bacillus subtilis-infecting bacteriophages in soybean-based fermented foods and its detrimental effects on the process and quality of Cheonggukjang. Food Microbiol 76:196–203
Ghosh K, Senevirathne A, Kang H, Hyun W, Kim J, Kim KP (2018) Complete nucleotide sequence analysis of a novel Bacillus subtilis-infecting bacteriophage BSP10 and its effect on poly-gamma-glutamic acid degradation. Viruses 10:240
Gu W, Jackman JE, Lohan AJ, Gray MW, Phizicky EM (2003) tRNAHis maturation: An essential yeast protein catalyzes addition of a guanine nucleotide to the 5′ end of tRNAHis. Genes Dev 17:2889–2901
Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704
Kimura K, Itoh Y (2003) Characterization of poly-γ-glutamate hydrolase encoded by a bacteriophage genome: possible role in phage infection of Bacillus subtilis encapsulated with poly-γ-glutamate. Appl Environ Microbiol 69:2491–2497
Laslett D, Canback B (2004) ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res 32:11–16
Lefkowitz EJ, Dempsey DM, Hendrickson RC, Orton RJ, Siddell SG, Smith DB (2018) Virus taxonomy: the database of the International Committee on Taxonomy of Viruses (ICTV). Nucleic Acids Res 46:D708–D717
Nagai T (2012) Bacteriophages of Bacillus subtilis (natto) and their contamination in natto factories. In: Kurtboke DI (ed) Bacteriophages. InTech, Rijeka, pp 95–110
Nameki N, Asahara H, Shimizu M, Okada N, Himeno H (1995) Identity elements of Saccharomyces cerevisiae tRNA His. Nucleic Acids Res 23:389–394
Rao BS, Maris EL, Jackman JE (2011) tRNA 5′-end repair activities of tRNA His guanylyltransferase (Thg1)-like proteins from bacteria and archaea. Nucleic Acids Res 39:1833–1842
Rombel IT, Sykes KF, Rayner S, Johnston SA (2002) ORF-FINDER: a vector for high-throughput gene identification. Gene 282:33–41
Stothard P, Wishart DS (2004) Circular genome visualization and exploration using CGView. Bioinformatics 21:537–539
Zafar N, Mazumder R, Seto D (2002) CoreGenes: a computational tool for identifying and cataloging” core” genes in a set of small genomes. BMC Bioinform 3:12
Acknowledgements
This work was supported by the National Research Foundation of Korea (Project no. NRF-2016R1A2B4006967) and by research funds of Chonbuk National University in 2018.
Funding
This study was funded by the National Research Foundation of Korea (Project no. NRF-2016R1A2B4006967) and by research funds of Chonbuk National University in 2018.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent
Not applicable for this study.
Additional information
Handling Editor: Chan-Shing Lin.
Nucleotide sequence accession number
The whole genome sequence of Bacillus phage BSP38 was deposited in GenBank under accession number MH606185 (http://www.ncbi.nlm.nih.gov/nuccore/MH606185).
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ghosh, K., Kim, KP. Complete nucleotide sequence analysis of a novel Bacillus subtilis-infecting phage, BSP38, possibly belonging to a new genus in the subfamily Spounavirinae. Arch Virol 164, 875–878 (2019). https://doi.org/10.1007/s00705-018-4110-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-018-4110-5