Abstract
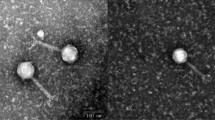
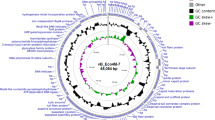
The genome sequence of a novel virulent bacteriophage, named “ C130_2”, that is morphologically a member of the family Myoviridae is reported. The 41,775-base-pair double-stranded DNA genome of C130_2 contains 59 ORFs but exhibits overall low sequence similarity to bacteriophage genomes for which sequences are publicly available. Phylogenetic analysis indicated that C130_2 represents a new phage type. C130_2 could be propagated well on enterohemorrhagic Escherichia coli (EHEC) O157:H7 and other pathogenic E. coli strains, as well as on strains of various Shigella species.


Similar content being viewed by others
References
Hagens S, Loessner MJ (2010) Bacteriophage for biocontrol of foodborne pathogens: calculations and considerations. Curr Pharm Biotechnol 11:58–68. https://doi.org/10.2174/138920110790725429
Abedon ST (2017) Bacteriophage clinical use as antibacterial “drugs”: utility and Precedent. Microbiol Spectr. https://doi.org/10.1128/microbiolspec.BAD-0003-2016
Hong Y, Pan Y, Harman NJ, Ebner PD (2014) Complete genome sequences of two Escherichia coli O157:H7 phages effective in limiting contamination of food products. Genome Announc. https://doi.org/10.1128/genomeA.00519-14
Sváb D, Falgenhauer L, Rohde M et al (2018) Identification and characterization of T5-like bacteriophages representing two novel subgroups from food products. Front Microbiol 9:202. https://doi.org/10.3389/fmicb.2018.00202
Kim M, Heu S, Ryu S (2014) Complete genome sequence of enterobacteria phage 4MG, a new member of the subgroup “PVP-SE1-like phage” of the “rV5-like viruses”. Arch Virol 159:3137–3140. https://doi.org/10.1007/s00705-014-2140-1
Liu H, Niu YD, Meng R et al (2015) Control of Escherichia coli O157 on beef at 37, 22 and 4 °C by T5-, T1-, T4-and O1-like bacteriophages. Food Microbiol 51:69–73. https://doi.org/10.1016/j.fm.2015.05.001
Lee H, Ku H-J, Lee D-H et al (2016) Characterization and genomic study of the novel bacteriophage HY01 infecting both Escherichia coli O157:H7 and Shigella flexneri: potential as a biocontrol agent in food. PLoS One 11:e0168985. https://doi.org/10.1371/journal.pone.0168985
Ramirez K, Cazarez-Montoya C, Lopez-Moreno HS, Castro-Del Campo N (2018) Bacteriophage cocktail for biocontrol of Escherichia coli O157:H7: stability and potential allergenicity study. PLoS One 13:e0195023. https://doi.org/10.1371/journal.pone.0195023
Wang C, Chen Q, Zhang C et al (2017) Characterization of a broad host-spectrum virulent Salmonella bacteriophage fmb-p1 and its application on duck meat. Virus Res 236:14–23. https://doi.org/10.1016/j.virusres.2017.05.001
Soffer N, Woolston J, Li M et al (2017) Bacteriophage preparation lytic for Shigella significantly reduces Shigella sonnei contamination in various foods. PLoS One 12:e0175256. https://doi.org/10.1371/journal.pone.0175256
Nagy B, Szmolka A, Smole Možina S et al (2015) Virulence and antimicrobial resistance determinants of verotoxigenic Escherichia coli (VTEC) and of multidrug-resistant E. coli from foods of animal origin illegally imported to the EU by flight passengers. Int J Food Microbiol 209:52–59. https://doi.org/10.1016/j.ijfoodmicro.2015.06.026
Sambrook J, Maniatis T, Fritsch EF, Laboratory CSH (1987) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Tóth I, Sváb D, Bálint B et al (2016) Comparative analysis of the Shiga toxin converting bacteriophage first detected in Shigella sonnei. Infect Genet Evol J Mol Epidemiol Evol Genet Infect Dis 37:150–157. https://doi.org/10.1016/j.meegid.2015.11.022
Bankevich A, Nurk S, Antipov D et al (2012) SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. https://doi.org/10.1089/cmb.2012.0021
Overbeek R, Olson R, Pusch GD et al (2014) The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res 42:D206–D214. https://doi.org/10.1093/nar/gkt1226
Lowe TM, Chan PP (2016) tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res 44:W54–W57. https://doi.org/10.1093/nar/gkw413
Meier-Kolthoff JP, Göker M (2017) VICTOR: genome-based phylogeny and classification of prokaryotic viruses. Bioinformatics 33:3396–3404. https://doi.org/10.1093/bioinformatics/btx440
Hua Y, An X, Pei G et al (2014) Characterization of the morphology and genome of an Escherichia coli podovirus. Arch Virol 159:3249–3256. https://doi.org/10.1007/s00705-014-2189-x
Sullivan MJ, Petty NK, Beatson SA (2011) Easyfig: a genome comparison visualizer. Bioinforma Oxf Engl 27:1009–1010. https://doi.org/10.1093/bioinformatics/btr039
Anderson M, Sansonetti PJ, Marteyn BS (2016) Shigella diversity and changing landscape: insights for the twenty-first century. Front Cell Infect Microbiol 6:45. https://doi.org/10.3389/fcimb.2016.00045
Nüesch-Inderbinen M, Heini N, Zurfluh K et al (2016) Shigella antimicrobial drug resistance mechanisms, 2004–2014. Emerg Infect Dis 22:1083–1085. https://doi.org/10.3201/eid2206.152088
Blattner FR, Plunkett III G, Bloch CA et al (1997) The complete genome sequence of Escherichia coli K-12. Science 277:1453–1462. https://doi.org/10.1126/science.277.5331.1453
Hayashi T, Makino K, Ohnishi M et al (2001) Complete genome sequence of enterohemorrhagic Eschelichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res 8:11–22. https://doi.org/10.1093/dnares/8.1.11
Perna NT, Plunkett G, Burland V et al (2001) Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529–533. https://doi.org/10.1038/35054089
Marchès O, Ledger TN, Boury M et al (2003) Enteropathogenic and enterohaemorrhagic Escherichia coli deliver a novel effector called Cif, which blocks cell cycle G2/M transition. Mol Microbiol 50:1553–1567
Sváb D, Bálint B, Maróti G, Tóth I (2016) Cytolethal distending toxin producing Escherichia coli O157:H43 strain T22 represents a novel evolutionary lineage within the O157 serogroup. Infect Genet Evol J Mol Epidemiol Evol Genet Infect Dis. https://doi.org/10.1016/j.meegid.2016.11.003
Iguchi A, Thomson NR, Ogura Y et al (2009) Complete genome sequence and comparative genome analysis of enteropathogenic Escherichia coli O127:H6 strain E2348/69. J Bacteriol 191:347–354. https://doi.org/10.1128/JB.01238-08
Hochhut B, Wilde C, Balling G et al (2006) Role of pathogenicity island-associated integrases in the genome plasticity of uropathogenic Escherichia coli strain 536. Mol Microbiol 61:584–595. https://doi.org/10.1111/j.1365-2958.2006.05255.x
Moriel DG, Bertoldi I, Spagnuolo A et al (2010) Identification of protective and broadly conserved vaccine antigens from the genome of extraintestinal pathogenic Escherichia coli. Proc Natl Acad Sci USA 107:9072–9077. https://doi.org/10.1073/pnas.0915077107
Tóth I, Nougayrède J-P, Dobrindt U et al (2009) Cytolethal distending toxin type I and type IV genes are framed with lambdoid prophage genes in extraintestinal pathogenic Escherichia coli. Infect Immun 77:492–500. https://doi.org/10.1128/IAI.00962-08
Allué-Guardia A, García-Aljaro C, Muniesa M (2011) Bacteriophage-encoding cytolethal distending toxin type V gene induced from nonclinical Escherichia coli isolates. Infect Immun 79:3262–3272. https://doi.org/10.1128/IAI.05071-11
Sváb D, Bálint B, Vásárhelyi B et al (2017) Comparative genomic and phylogenetic analysis of a Shiga toxin producing Shigella sonnei (STSS) strain. Front Cell Infect Microbiol 7:229. https://doi.org/10.3389/fcimb.2017.00229
Petty NK, Bulgin R, Crepin VF et al (2010) The Citrobacter rodentium genome sequence reveals convergent evolution with human pathogenic Escherichia coli. J Bacteriol 192:525–538. https://doi.org/10.1128/JB.01144-09
Acknowledgements
This study was supported by the Hungarian National Research, Development and Innovative Office (NKFIH, Grant 124335), 7th EU Framework Programme PROMISE project (Project No. 265877) and by grants to the German Center of Infection Research (DZIF, site Giessen-Marburg-Langen), through the German Federal Ministry of Education and Research, Grant # TI06.001 and 8032808811 to TC. We thank Christina Gerstmann for excellent technical assistance. Domonkos Sváb was supported by the János Bolyai Research Scholarship of the Hungarian Academy of Sciences.
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling Editor: Tim Skern.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Table 1
Host spectrum and efficiency of plating (EOP) of bacteriophage C130_2. EOP values are given relative to the titre measured on E. coli K-12 MG1655 strain (XLSX 12 kb)
Supplementary Table 2
List of ORFs of phage C130_2 with assigned functions and protein homology searches. A PROSITE search was performed including motifs with high probability occurrences. A UniProt search was performed with narrowing down to viral proteins (XLSX 471 kb)
Supplementary Table 3
List of ORFs of phages C130_2, IME_EC2 and vB_KpnS_IME279, with corresponding ORFs above 75% similarity highlighted in blue (XLSX 14 kb)
Supplementary Fig. 1
BLAST-based comparison of the whole genomes of bacteriophages C130_2, IME_EC2 and vB_KpnS_IME279. Orange arrows represent genes, numbers on C130_2 genes correspond to ORF numbers in Supplementary Tables 2 and 3. Regions showing > 50% similarity are interconnected with grey lines (TIFF 1220 kb)
Rights and permissions
About this article
Cite this article
Sváb, D., Falgenhauer, L., Rohde, M. et al. Complete genome sequence of C130_2, a novel myovirus infecting pathogenic Escherichia coli and Shigella strains. Arch Virol 164, 321–324 (2019). https://doi.org/10.1007/s00705-018-4042-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-018-4042-0