Abstract
The genomes of two new lytic phages of Corynebacterium glutamicum ATCC 13032, φ673 and φ674, were sequenced and annotated (GenBank: MG324353, MG324354). Electron microscopy studies of both virions revealed that taxonomically they belong to the Siphoviridae family and have a polyhedral head with a width of 50 nm and a non-contractile tail with a length of 250 nm. The genomes of φ673 and φ674 consist of linear double-stranded DNA molecules with lengths of 44,530 bp (G+C = 51.1%) and 43,193 bp (G+C = 50.7%) and identical, protruding, cohesive 3’ ends 13 nt in length. The level of identity between the φ673 and φ674 genomes is 85.2%. Two major structural proteins of each virion were separated via SDS-PAGE and identified using peptide mass fingerprinting. Based on bioinformatic analysis, 56 and 54 ORFs were predicted for φ673 and φ674, respectively. Only 20 of the putative gene products of φ673 and 20 of φ674 could be assigned to known functions. Both genomes were divided into functional modules. Nine putative promoters in the φ673 genome and eight in the φ674 genome were predicted. One bidirectional Rho-independent transcription terminator was identified and experimentally confirmed in each phage genome.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Corynebacterium glutamicum is a nonpathogenic, gram-positive bacterium that is widely used for the industrial production of a broad range of substances, including amino acids and proteins [1]. In many cases, phages are responsible for the lysis of commercially interesting strains during fermentation, which leads to financial losses in the biotechnology industry. Many corynephages have been isolated, but only a few of them have been completely sequenced [2,3,4]. In the present study, the genomes of φ673 and φ674, two newly identified lytic phages of C. glutamicum ATCC 13032, were sequenced and annotated. The phages φ673 and φ674 were obtained from VKPM (the Russian National Collection of Industrial Microorganisms at the Institute of Genetics and Selection of Industrial Microorganisms, Moscow). Four genes associated with sensitivity to φ674 were identified in the C. glutamicum ATCC 13032 genome and could be useful for the construction of phage-resistant strains [5]. The newly constructed cosmid based on cos-sites of φ674 could be helpful for improving genetic tools for C. glutamicum, particularly with respect to the non-specific transduction of DNA fragments between C. glutamicum ATCC 13032 strains; such transduction has been reported for other phage-host systems [6, 7].
Results and discussion
Phages φ673 and φ674 were propagated on C. glutamicum ATCC 13032 and purified via centrifugation in a CsCl gradient as previously described [8].
Transmission electron microscopy studies of these two phages revealed that their virions belong to the Siphoviridae family. Both virions had a polyhedral head with a width of 50 nm and a long non-contractile tail with a length of 250 nm and a diameter of 11 nm (Fig. 1a, b). The putative gene products (gp) gpφ67314 and gpφ67414 were assigned to the tail tape measure protein (TMP). For both phages, the relationship between the observed tail length (~ 250 nm) and TMP size (1,577 aa for φ673 and 1,572 aa for φ674), which involved a ratio of 0.159 nm/aa, was reasonable [9].
(a, b) Electron micrograph of φ673 and φ674 phages. Bar, 50 nm. (d) SDS-PAGE analysis of φ673 and φ674 structural proteins. Molecular weight markers (lane I). Protein profile of φ673 (lane II), φ674 (lane III). (d) Two major proteins bands from the φ673 and φ674 phages underwent peptide mass fingerprinting analysis; (c, f) the corresponding predicted amino acid sequences (not highlighted) and the aa sequences detected in the analysis (highlighted) are shown
Purified genomic DNA from both phages was sequenced using Illumina technology at Evrogen (Moscow, Russia, http://www.evrogen.ru). Sequences of cos-sites were determined in run-off experiments and were compared with the nucleotide sequences of the ligated phage ends.
Two online bioinformatic programs, Glimmer3 (https://www.ncbi.nlm.nih.gov/genomes/MICROBES/glimmer_3.cgi) and GeneMark S (http://exon.biology.gatech.edu/), were used to search for ORFs. InterPro (http://www.ebi.ac.uk/interpro/) was used to improve the initial annotation of predicted proteins. Putative promoters were searched using phiSITE’s PromoterHunter (with parameters for “-10” and “-35” [Supplementary Fig. 1]) (http://www.phisite.org/main/index.php?nav=tools&nav_sel=hunter). Bi-directional, while rho-independent transcription terminators were identified using ARNold: finding terminators (http://rna.igmors.u-psud.fr/toolbox/arnold/index.php).
The φ673 and φ674 genomes consist of linear double-stranded DNA molecules with lengths of 44,530 bp (G+C = 51.1%) and 43,193 bp (G+C = 50.7%), respectively, and share identical, protruding, cohesive 3’ ends 13 nt in length (AGAAGGGGGCGGA-3’). A cosmid vector for molecular cloning has been constructed on the basis of the phage φ674 cos-site, and the functionality of the cos-site was experimentally confirmed (unpublished results). Based on bioinformatics analysis, 56 and 54 ORFs were identified in the φ673 and φ674 genomes, respectively. These ORFs cover approximately 97% and 96% of the entire φ673 and φ674 genomes, respectively. Only 20 gene products (gps) from each phage could be assigned to known biological functions (Supplementary Table 1, 2); the other 17 and 16 gp(s) exhibiting homology to hypothetical proteins, while 19 and 18 ORFs present in φ673 and φ674, respectively, had no homologues in the databases. No tRNA genes were identified in either phage genomes.
Nine and eight putative promoters were predicted in the φ673 and φ674 genomes, respectively (Supplementary Table 3, 4). One bidirectional, rho-independent transcription terminator was identified in each phage genome (Supplementary Fig. 2) and experimentally confirmed (unpublished result).
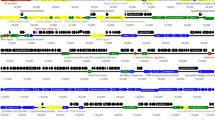
Based on homology to known phage proteins, functional domains, and mutual arrangement, putative functions were assigned to products of 20 of the predicted ORFs in each phage (Supplementary Table 1, 2). For each phage, the entire genome was divided into four functional modules (Fig. 2). The DNA packaging module includes small (gpφ6731 and gpφ6741) and large (gpφ6732 and gpφ6742) terminase subunits and a portal protein (gpφ6733 and gpφ6743). A head maturation protease (gpφ6734 and gpφ6744), major capsid and tail proteins (gpφ6735 and gpφ6745 and gpφ67311 and gpφ67411), head-to-tail connectors (gpφ6737, 8, 9 and gpφ6747, 8, 9), a tail assembly chaperone (gpφ67312 and gpφ67412), a tail TMP (gpφ67314 and gpφ67414), a tail protein (gpφ67316 and gpφ67416) and a tail fiber protein (gpφ67319, 21 and gpφ67419) could be predicted in the structural components and assembly module. Two major structural proteins for each virion, the major capsid (gpφ6735 and gpφ6745) and tail (gpφ67311 and gpφ67411) proteins, were detected via SDS-PAGE and identified via trypsin-based peptide mass fingerprinting (PMF) using an Ultraflex II LC-MALDI-TOF/TOF (Bruker) in accordance with a previously described procedure [10] (Fig. 1c, d, e). Furthermore, elimination of an N-terminal Met residue retained in trypsin-digested peptides from gpφ67311 and gpφ67411 confirmed the predicted N-terminal processing rule [11] (Fig. 1c, e).
Genomic organization of φ673 (a) and φ674 (b) phages. ORFs are numbered consecutively from left to right and are indicated by arrows in the direction of transcription. ORFs encoding proteins assigned to known biological functions are depicted as dark arrows while those without known function are light. ORFs, joined by braces, are indicated to represent the proposed functional modules within the phage genomes. Promoter positions and directions are indicated by thin arrows, while intrinsic terminators are indicated by boxes
A homolog of a known enzyme, lysozyme-like protein (gpφ67322 and gpφ67420), was predicted in the host lysis module. The replication/recombination/metabolism module also contained homologs to known proteins, including helicase (gpφ67343 and gpφ67440), the DNA replication protein RepA primase/helicase (gpφ67345 and gpφ67442), DNA polymerase I (gpφ673 46 and gpφ674 44) and HNH endonuclease (gpφ67333 and gpφ67430, 31, 43). One transcriptional regulator, gpφ673 27, was identified. Interestingly, a putative intein was identified in the helicase encoded in ORF 43 for φ673, in contrast to the helicase encoded in ORF 40 for φ674, which exhibited no inteins. It has previously been reported that the Corynebacterium phage P1201 contains inteins [3].
Significant similarity throughout the genome was observed between the two newly sequenced and annotated lytic corynephages, φ673 and φ674, which exhibited approximately 85.2% identity. A bioinformatics search revealed that both phage genomes had high similarity to the genome of the corynephage BFK20 [2], with approximately 55% identity. Multiple genome alignment was constructed with Mauve (ver. 2.2.0) (Supplementary Fig. 3).
Besides C. glutamicum ATCC 13032, the host strain for both φ673 and φ674 phages, MB001 (prophage-free variant of C. glu ATCC 13032) was also infected by both phages. Another tested wild-type strain Brevibacterium lactofermentum AJ1511 was not lysed by either of the two phages.
We identified four C. glutamicum ATCC13032 genes, responsible for phage φ674 sensitivity (unpublished results). Two of these genes encoded glycosyltransferases; these proteins are bacterial sugar transferases involved in lipopolysaccharide synthesis. The third gene is annotated as a gene encoding a putative secreted protein. The fourth gene encodes a nucleotidyltransferase/DNA polymerase involved in DNA repair that is a DNA polymerase IV homolog. We hypothesized that these glycosyltransferases participate in the synthesis of a φ674 phage receptor containing an unknown sugar component in its structure [12].
In summary, the genomes of the φ673 and φ674 phages are significantly different from existing corynephage genomes available in databases; therefore, the sequences of these complete phage genomes were deposited for the first time in GenBank under accession numbers: MG324353, MG324354.
References
Becker J, Wittmann C (2012) Bio-based production of chemicals, materials and fuels—Corynebacterium glutamicum as versatile cell factory. Curr Opin Biotechnol 23:631–640. https://doi.org/10.1016/j.copbio.2011.11.012
Bukovska G, Klucar L, Vlcek C, Adamovic J, Turna J, Timko J (2006) Complete nucleotide sequence and genome analysis of bacteriophage BFK20—a lytic phage of the industrial producer Brevibacterium flavum. Virology 348:57–71. https://doi.org/10.1016/j.virol.2005.12.010
Chen CL, Pan TY, Kan SC et al (2008) Genome sequence of the lytic bacteriophage P1201 from Corynebacterium glutamicum NCHU 87078: evolutionary relationships to phages from Corynebacterineae. Virology 378:226–232. https://doi.org/10.1016/j.virol.2008.05.027
Lobanova JS, Gak ER, Andreeva IG, Rybak KV, Krylov AA, Mashko SV (2017) Complete nucleotide sequence and annotation of the temperate corynephage ϕ16 genome. Arch Virol 162(8):2489–2492. https://doi.org/10.1007/s00705-017-3383-4
Labrie SJ, Samson JE, Moineau S (2010) Bacteriophage resistance mechanisms. Nat Rev Microbiol 8(5):317–327. https://doi.org/10.1038/nrmicro2315
Thomason LC, Costantino N, Court DL, Thomason LC, Costantino N, Court DL (2007) Genome manipulation by P1 transduction. Curr Protoc Mol Biol Chapter 1:Unit 1.17. https://doi.org/10.1002/0471142727.mb0117s79
Van Kessel JC, Marinelli LJ, Hatfull GF (2008) Recombineering Mycobacteria and their phages. Nat Rev Microbiol 6(11):851–857. https://doi.org/10.1038/nrmicro2014
Boulanger P (2009) Purification of bacteriophages and SDS-PAGE analysis of phage structural proteins from host particles. Methods Mol Biol 502:227–238. https://doi.org/10.1007/978-1-60327-565-1_13
Abuladze NK, Gingery M, Tsai J, Eiserling FA (1994) Tail length determination in bacteriophage T4. Virology 199(2):301–310. https://doi.org/10.1006/viro.1994.1128
Govorun VM, Moshkovskii SA, Tikhonova OV et al (2003) Comparative analysis of proteome maps of Helicobacter pylori clinical isolates. Biochemistry (Mosc) 68:42–49
Eggeling L, Bott M (2005) Handbook of Corynebacterium glutamicum. CRC Press, Boca Raton
Lovering AL, de Castro LH, Lim D, Strynadka NCJ (2007) Structural insight into the transglycosylation step of bacterial cell-wall biosynthesis. Science 315(5817):1402–1405. https://doi.org/10.1126/science.1136611
Acknowledgements
The authors are grateful to Dr. Alexander A. Krylov for helpful discussions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
There is no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Handling Editor: T. K. Frey.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Yomantas, Y.A.V., Abalakina, E.G., Lobanova, J.S. et al. Complete nucleotide sequences and annotations of φ673 and φ674, two newly characterised lytic phages of Corynebacterium glutamicum ATCC 13032. Arch Virol 163, 2565–2568 (2018). https://doi.org/10.1007/s00705-018-3867-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-018-3867-x