Abstract
Ustilaginoidea virens is the causal agent of a serious disease of rice. Here, we report the presence of five dsRNA bands ranging from about 1.2, 1.5, 1.7, and 1.8 to 5.6 kb in an isolate of this fungus from China and the complete sequence of the largest dsRNA segment, putatively representing the genome of a novel virus, designated as Ustilaginoidea virens RNA virus 1 (UvRV1), UvRV1, which has a genome length of 5567 bp and has two consecutive open reading frames (ORFs) with a five-nucleotide overlap. Phylogenetic analysis showed that UvRV1 belongs to the genus of Victorivirus in the family Totiviridae.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mycoviruses with double-stranded RNA (dsRNA) genomes have been described in a wide range of fungi covering all four phyla of the true fungi: Chytridiomycota, Zygomycota, Ascomycota, and Basidiomycota. Mycoviruses are classified into 11 families, and there are many that have not yet been assigned to a family [1]. Members of five of these 11 families, Totiviridae, Partitiviridae, Chrysoviridae, Reoviridae, and Megabirnaviridae, have double-stranded (ds) RNA genomes encapsidated in rigid virus particles. The type of dsRNA segments observed in fungal isolates can be quite diverse, even in the same fungal species [2]. This diversity can be seen in the number and size of the dsRNA segments detected in a fungal strain, and it could indicate multipartite viral genomes, mixed infections, or even defective products of virus replication [3].
Rice false smut is a worldwide fungal disease, caused by U. virens (teleomorph: Villosiclava virens) [4–6]. The disease was first reported from the Tirunelveli district of the Tamil Nadu state of India [6]. In recent years, rice false smut has become more epidemic in areas of Asia, America, and Europe, where new hybrid varieties were widely planted, highly efficient cultivation methods were adopted, and chemical fertilizers were extensively used [7–11]. A characteristic trait of rice false smut is the formation of ball-like colonies in spikelets, which begin to appear 10 to 15 days after rice anthesis. The disease significantly reduces grain quality and yield of rice [8]. Yield losses caused by rice false smut disease have been estimated to vary between 0.2 % and 49 % depending on the infection severity and rice variety [11, 12].
At present, there is no record of a double-stranded RNA virus infecting U. virens. In this study, we report the presence of dsRNAs in a sample of U. virens isolated from rice in China.
Provenance of the virus material
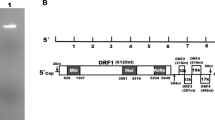
Ustilaginoidea virens strain JYH-ZT, used in this study, was originally isolated from rice cultivated in Hubei, China. This strain was maintained on potato sucrose agar (PSA) plates. For extraction and purification of dsRNAs, mycelia were grown on potato sucrose broth (PSB) with shaking (150 rpm) at 28 °C. dsRNA was extracted from fungal mycelia using CF-11 cellulose (Sigma, St. Louis, MO, USA) column chromatography as described previously [13, 14]. To remove contaminating DNA and ssRNA, we treated the dsRNA sample with RNase-free DNaseI (TAKALA, Dalian, China; RNase-free DNaseI) and S1 nuclease (TAKALA Dalian, China; S1 nuclease) at 37 °C for 30 min. The dsRNA sample was analyzed by 1 % (w/v) agarose gel electrophoresis containing TAE buffer (40 mM Tris-acetate, 2 mM EDTA, pH 8.1) and 500 ng/ml ethidium bromide, and the largest dsRNA fragment, called dsRNA1 (Fig. 1), was extracted from the gel, denatured and used for reverse transcription and PCR amplification. A random-primer amplification method [15] was used to obtain the full-length sequence of dsRNA1. The ends of the molecule were cloned using an improved method [16]. Sequence analysis, alignment, and phylogenetic analysis were performed using DNAMAN, the COBALT web server (http://www.ncbi.nlm.nih.gov/tools/cobalt/cobalt.cgi?link_loc=BlastHomeLink), and the program PhyML 3.0 [17], respectively. The other four dsRNA elements present in the U. virens strain JYH-ZT were termed dsRNA2 (1.8 kb), dsRNA3 (1.7 kb), dsRNA4 (1.5 kb), and dsRNA5 (1.2 kb), respectively, according to their sizes (Fig. 1). Their sequences and biological functions are under further investigation.
Agarose gel electrophoresis of dsRNA extracted from mycelia of Ustilaginoidea virens strain JYH-ZT. The dsRNA preparations were treated with RNase-free DNaseI and S1 nuclease, fractionated on 1.0 % agarose gels and stained with ethidium bromide. Lane M, Trans2 k PlusII DNA Marker (TranGen Biotech, Beijing), used as a size marker; sizes in kb indicated at the left. Lane I, five dsRNA elements from U. virens are displayed
Sequence properties
The complete sequence of dsRNA1 was 5567 bp and had a GC content of 57 %. Sequence analysis showed that dsRNA1 had two consecutive open reading frames (ORFs) (Fig. 2). ORF1 had a length of 2175 bp and encoded a 725-amino-acid protein (76.189 kDa). ORF2 was 2478 bp in length and encoded an 826-amino-acid protein (91.629 kDa). For the gene sequences, the stop codon of the first gene and the start codon of the following gene overlapped in the coupled termination-reinitiation model of protein expression [18, 19]. Based on this model, we predicted that dsRNA1 contained two ORFs that were demarcated by a pentanucleotide, UAAUG, which constituted the stop codon of ORF1 and the start codon of ORF2. This five-nucleotide overlap has been described in other totiviruses, such as Aspergillus foetidus virus slow-1 [20] and Beauveria bassiana RNA virus 1 (EMBL/GenBank accession number: CCC42235). The amino acid sequence deduced from ORF1 of dsRNA1 showed a high level of similarity to those of the capsid proteins (CP) of viruses of the family Totiviridae, particularly to that of Beauveria bassiana RNA virus 1 (BbRV1; 61 % identity). The C-terminus of this putative CP had an Ala/Gly/Pro-rich region, which occurs in mycoviruses of the genus Victorivirus [18]. The protein encoded by ORF2 contained conserved motifs characteristic of viral RNA-dependent RNA polymerases (RdRps) and appeared most similar (54 % identity) to an ortholog encoded by BbRV1. The 5’ untranslated region (UTR) in ORF1 had 394 bp and started with a CTTTG sequence, which was the same as the CTTTG motif present in the genome of type II Trichomonas vaginalis virus 2 (TVV2) [21], and similar to the CTTAA motif present in the genome of type I T. vaginalis virus 1 (TVV1) [22]. The GC content of this region was 52 %. The 3’ UTR in ORF2 had a length of 509 bp and a GC content of 58 %.
A phylogenetic analysis based on the complete amino acid sequence of putative RdRps encoded by dsRNA1 and totiviruses showed close relationships between dsRNA1 and members of the genus Victorivirus (Fig. 3). The phylogenetic analysis showed that this dsRNA element represented a new member of the genus Victorivirus, which was called UvRV1. A sequence comparison analysis indicated that the eight conserved motifs of the RdRps sequences of the dsRNA viruses in filamentous ascomycetous fungi [23] were present in this putative RdRp of UvRV1 (Supplementary Fig. 1). Finally, based upon our phylogenetic analysis and the Victorivirus species demarcation criteria established by the International Committee on Taxonomy of Viruses, which states that the amino acid sequence identity in pairwise comparisons of CP or RdRp gene products between members of different species is no more than 60 % [24], UvRV1 should be considered a new member of the genus Victorivirus. The complete genome sequence has been deposited in the EMBL nucleotide sequence database with the accession number JX524563.
Phylogenetic tree of viruses of the family Totiviridae based on RdRp amino acid sequences. The tree was constructed by the maximum-likelihood (ML) method using the program PhyML 3.0 and the LG model of amino acid substitution. The reliability of internal branches was evaluated based on SH-aLRT support. Abbreviations and GenBank accession numbers are as follows: Aspergillus foetidus virus slow-1 (AfV-S1; HE588147); Sphaeropsis sapinea RNA virus 1 (SsRV1; NP047588); Beauveria bassiana RNA virus 1 (BbRV1; CCC42235); Botryotinia fuckeliana totivirus 1 (BfTV1; YP001109580); Tolypocladium cylindrosporum virus 1 (TcV1; YP004089630); Helminthosporium victoriae virus 190S (HvV190S; NP61967); Magnaporthe oryzae virus 1 (MoV1; YP122352); Helicobasidium mompa totivirus 1-17 (HmTV1-17; NP898833); Aspergillus mycovirus 178 (AMV178; ABX7995); Gremmeniella abietina RNA virus L2 (GaRV-L2; YP044807); Gremmeniella abietina RNA virus L1 (GaRV-L1; AAK11656); Magnaporthe oryzae virus 2 (MoV2; YP001649206); Epichloe festucae virus 1 (EfV1; CAK02788); Coniothyrium minitans RNA virus (CmRV; YP392467); Sphaeropsis sapinea RNA virus 2 (SsRV2; NP047560); Eimeria brunetti RNA virus 1 (EbRV1; NP108651); Leishmania RNA virus 1–1, (LRV1-1; NP041191); Leishmania RNA virus 1–4 (LRV1-4; NP619653); Leishmania RNA virus 2–1 (LRV2-1; NP043465); Trichomonas vaginalis virus 1 (TvV1; AAA62868); Trichomonas vaginalis virus 2 (TvV2; AAF29445); Trichomonas vaginalis virus (TvV3; NP659390); Saccharomyces cerevisiae virus L-A (L1) (ScV-L-A; NP620495); Saccharomyces cerevisiae virus L-BC (ScV-L-BC; NP042581); Ustilago maydis virus H1 (UmV-H1; U01059); Giardia lamblia virus (GLV; NP620070); Armigeres subalbatus virus SaX06-AK20 (AsTV; ACH85916); penaeid shrimp infectious myonecrosis virus (IMNV; AAT67231); Ophiostoma minus totivirus (OmV; CAJ34336.1); Chalara elegans RNA virus 1 (CeRV1; AY561500); Drosophila melanogaster totivirus SW-2009a (DTV; GQ342961); Omono River virus, strain AK4 (OMRV-AK4; AB555544); Omono River virus, strain AK4 (OMRV-Y61; AB555545); Ustilaginoidea virens RNA virus 1 (UvRV1)
References
Ghabrial SA, Suzuki N (2009) Viruses of plant pathogenic fungi. Annu Rev Phytopathol 47:353–384
Pearson MN, Beever RE, Boine B, Arthur K (2009) Mycoviruses of filamentous fungi and their relevance to plant pathology. Mole Plant Pathol 10(1):115–128
Ghabrial S, Suzuki N (2008) Fungal viruses. Encycl Virol 2:284–291
Padwick GW (1950) Manual of rice diseases. Commonwealth Mycological Institute, Kew, UK
Tanaka E, Ashizawa T, Sonoda R, Tanaka C (2008) Villosiclava virens gen. nov., comb. nov., the teleomorph of Ustilaginoidea virens, the causal agent of rice false smut. Mycotaxon 106:491–501
Cooke M (1878) Some extra-European fungi. Grevillea 7:13–15
Yaegashi H, Fujita Y, Sonoda R (1989) Severe outbreak of false smut of rice in 1988. Plant Prot 43(6):311–314
Osada S (1995) Effect of false smut occurrence on yield and quality of rice. Annual Report of the Society of Plant Protection of North Japan
Rush M, Shahjahan A, Jones J, Groth D (2000) Outbreak of false smut of rice in Louisiana. Plant Dis 84(1):100
Brooks SA, Anders MM, Yeater KM (2009) Effect of cultural management practices on the severity of false smut and kernel smut of rice. Plant Dis 93(11):1202–1208
Ladhalakshmi D, Laha G, Singh R, Karthikeyan A, Mangrauthia S, Sundaram R, Thukkaiyannan P, Viraktamath B (2012) Isolation and characterization of Ustilaginoidea virens and survey of false smut disease of rice in India. Phytoparasitica 40:171–176
Dodan D, Singh R (1996) False smut of rice: present status. Agric Rev Agric Res Commun Cent India 17:227–240
Dodds JA, Bar-Joseph M (1983) Double-stranded RNA from plants infected with closteroviruses. Phytopathology 73(3):419–423
Hunst PL, Latterell FM, Rossi AE (1986) Variation in double-stranded RNA from isolates of Pyricularia oryzae. Phytopathology 76(7):674–678
Froussard P (1993) rPCR: a powerful tool for random amplification of whole RNA sequences. Genome Res 2(3):185–190
Potgieter AC, Page NA, Liebenberg J, Wright IM, Landt O, van Dijk AA (2009) Improved strategies for sequence-independent amplification and sequencing of viral double-stranded RNA genomes. J Gen Virol 90(6):1423–1432
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59(3):307–321
Ghabrial SA, Nibert ML (2009) Victorivirus, a new genus of fungal viruses in the family Totiviridae. Arch Virol 154(2):373–379
Li H, Havens WM, Nibert ML, Ghabrial SA (2011) RNA sequence determinants of a coupled termination-reinitiation strategy for downstream open reading frame translation in Helminthosporium victoriae virus 190S and other Victoriviruses (Family Totiviridae). J Virol 85(14):7343–7352
Kozlakidis Z, Herrero N, Coutts RHA (2012) The complete nucleotide sequence of a totivirus from Aspergillus foetidus. Arch Virol 158(1):263–266
Bessarab IN, Liu HW, Ip CF, Tai JH (2000) The complete cDNA sequence of a type II Trichomonas vaginalis virus. Virology 267(2):350–359
Tai JH, Ip CF (1995) The cDNA sequence of Trichomonas vaginalis virus-T1 double-stranded RNA. Virology 206(1):773–776
Ghabrial SA (1998) Origin, adaptation and evolutionary pathways of fungal viruses. Virus Genes 16(1):119–131
Wickner RB, Ghabrial SA, Nibert ML, Patterson JL and Wang CC (2011) Family Totiviridae. In: King AMQ, Adams MJ, Carstens EB, and Lefkowitz EJ (eds) Virus Taxonomy, 9th ICTV Report, Elsevier Inc., Amsterdam, pp 639–650
Acknowledgments
This work was supported by the National Major Project for Crop Breeding (2011ZX08003-001) and the China Scholarship Council.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
705_2013_1615_MOESM1_ESM.pdf
Supplementary material 1: The eight conserved motifs of the sequences of RdRps of U. virens RNA virus 1 (UvRV1). These conserved motifs were constructed using the COBALT web server (http://www.ncbi.nlm.nih.gov/tools/cobalt/cobalt.cgi?link_loc=BlastHomeLink). Abbreviations and GenBank accession numbers are as follows: Helminthosporium victoriae virus 190S (HvV190S; NP61967); Sphaeropsis sapinea RNA virus 1 (SsRV1; NP047588); Magnaporthe oryzae virus 1 (MoV1; YP122352); Saccharomyces cerevisiae virus L-A (L1) (ScV-L-A; NP620495); Ustilaginoidea virens RNA virus 1 (UvRV1). (PDF 44 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Zhang, T., Jiang, Y., Huang, J. et al. Complete genome sequence of a putative novel victorivirus from Ustilaginoidea virens . Arch Virol 158, 1403–1406 (2013). https://doi.org/10.1007/s00705-013-1615-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-013-1615-9