Abstract
Infection with pathogenic influenza viruses is associated with intense inflammatory disease. Here, we investigated the innate immune response in mice infected with H5N1 A/Vietnam/1203/04 and with reassortant human H1N1 A/Texas/36/91 viruses containing the virulence genes hemagglutinin (HA), neuraminidase (NA) and NS1 of the 1918 pandemic virus. Inclusion of the 1918 HA and NA glycoproteins rendered a seasonal H1N1 virus capable of inducing an exacerbated host innate immune response similar to that observed for highly pathogenic A/Vietnam/1203/04 virus. Infection with 1918 HA/NA:Tx/91 and A/Vietnam/1203/04 were associated with severe lung pathology, increased cytokine and chemokine production, and significant immune cell changes, including the presence of CD11b+Gr-1+ cells in the blood, lung and bone marrow. Significant differential gene expression in the lung included pathways for cell death, apoptosis, production and response to reactive oxygen radicals, as well as arginine and proline metabolism and chemokines associated with monocyte and neutrophil/granulocyte accumulation and/or activation. Arginase was produced in the lung of animals infected with A/Vietnam/1204. These results demonstrate that the innate immune cell response results in the accumulation of CD11b+Gr-1+ cells and products that have previously been shown to contribute to T cell suppression.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Worldwide, annual influenza epidemics result in approximately 3-5 million cases of severe disease and 250,000 to 500,000 deaths (WHO, Fact sheet on influenza, http://www.who.int/mediacentre/factsheets/fs211/en/index.html). Occasionally, an influenza virus emerges that causes great mortality over several countries; the most notable example is the virus that caused the 1918 pandemic. It is estimated that upwards of 30 % of the world’s population was infected, resulting in nearly 20 to 50 million deaths worldwide [1].
Until an outbreak in Hong Kong claimed six human lives in 1997 (A/Hong Kong/156/97), H5N1 avian influenza viruses were thought to be incapable of infecting humans directly. However, the initial H5N1 outbreak and more recent H5N1, H7N7, and H9N2 epizootics have revealed that avian influenza viruses can infect humans without prior adaptation and can cause significant morbidity and mortality in infected individuals. Since 2003, there have been at least 539 confirmed human cases of H5N1 influenza virus infections with 318 deaths (WHO, Confirmed cases of avian influenza (H5N1), http://www.who.int/csr/disease/avian_influenza/country/cases_table_2011_04_06/en/index.html).
The basis for the high morbidity and mortality associated with 1918 and H5N1 viruses has been the subject of much debate and investigation. Historical data reveal that in fatal cases, 1918 pandemic virus infection resulted in an acute aggressive bronchopneumonia with epithelial necrosis, hemorrhage, and/or edema, with death typically associated with secondary bacterial pneumonia [2]. H5N1 infection has been associated with high viral loads and an intense inflammatory response that results in immune cell population changes and severe disease characterized by fulminant pneumonia and multi-organ-system failure [3, 4].
The advent of plasmid-based reverse genetics has allowed the generation of recombinant influenza viruses containing genes from the 1918 virus entirely from cloned cDNAs [5–8]. A study by Tumpey et al. showed that the HA and NA genes of the 1918 virus in the background of the seasonal H1N1 virus A/Texas/36/91 (Tx/91) rendered the virus lethal in mice without prior adaptation [9]. Similarly, Pappas et al. demonstrated that individual exchange of most 1918 influenza virus genes with those of Tx/91 did not alter virulence; however, substitution of the hemagglutinin (HA), neuraminidase (NA) or polymerase subunit PB1 greatly affected pathogenicity of the single-gene reassortants in mice [10].
Studies using animal models to investigate mechanisms underlying severe influenza have implicated the innate immune system in complicating lung tissue recovery [11]. Infections in mice and non-human primate models with the fully reconstructed 1918 virus and highly pathogenic H5N1 human isolates have confirmed an exacerbated host immune response, including histological observations of severe lung pathology, excessive cytokine/chemokine production and significant immune cell population changes, most notably increased macrophages and neutrophils in the lung [11, 12].
In this study, we used a combination of cytometric bead array, immunophenotyping by flow cytometry, and Affymetrix microarrays methods to evaluate the cytokine/chemokine response, immune cell population changes, and differences in gene expression profiles in animals infected with viruses of differing virulence, namely the H5N1 isolate, A/Vietnam/1203/04, and viruses expressing HA and NA, or HA, NA, and NS1 of the 1918 pandemic influenza virus in the genetic background of the human H1N1 virus A/Texas/36/91. Our objective was to extensively characterize host factors/mechanisms that might contribute to differential innate immune cell responses during influenza virus infection. The data revealed that pathogenic virus infection resulted in altered cytokine and chemokine production, with differential expression of genes related to recruitment and activation of innate immune cells, and significant accumulation of CD11b+Gr-1+ in the blood, lung and bone marrow of infected animals.
Methods
Ethics statement
All experiments were conducted according to protocols reviewed and approved by the Battelle Institutional Biosafety Committee and the Institutional Animal Care and Use Committee of Battelle, which adhere to the National Institutes of Health guidelines for the care and use of laboratory animals. Animals infected with influenza virus were housed in an ABSL-3 enhanced facility.
Viruses
Viruses possessing the 1918 influenza virus HA and NA segments and HA, NA and NS1 protein in the genetic background of the human H1N1 virus A/Texas/36/91 were described previously [12]. Recombinant viruses were plaque purified, and the sequences of the virus segments were confirmed by reverse transcription PCR and sequencing. Stocks of 1918 HA/NA:Tx/91 and 1918 HA/NA/NS:Tx/91 were propagated on Madin-Darby canine kidney (MDCK) cells. A/Texas/36/91, propagated on MDCK cells, was from the laboratory of Dr. Adolfo García-Sastre at the Mount Sinai Medical Center (New York, New York). The highly pathogenic A/Vietnam/1203/04 virus was propagated in the allantoic cavity of 10-day-old hen eggs at 37 °C. The allantoic fluid from infected eggs was harvested and stored at -70 °C. The A/Vn virus used for challenge experiments was passed a single time in embryonated eggs prior to use. Virus titers were determined by plaque and TCID50 assays in MDCK cells and are expressed as plaque-forming units per milliliter (PFU/mL) and TCID50/mL, respectively. All viruses were stored at -70 °C until use.
Experimental design
Female BALB/c mice (5-6 weeks of age) were purchased from Charles Rivers Laboratories (Wilmington, MA). Mice were housed in polycarbonate filter cages inside a Biosafety Level 3 enhanced facility (BSL-3+). Food and water were provided ad libitum. To confirm pathogenicity of the 1918 HA/NA:Tx/91 and 1918 HA/NA/NS:Tx/91 virus stocks, groups of 3-7 mice were anesthetized with ketamine and xylazine and challenged via the intranasal route (IN) with 1 × 106 PFU of Tx/91, 1918 HA/NA:Tx/91, or 1918 HA/NA/NS:Tx/91. For comparison, groups of mice were also challenged with 1.5 × 103 TCID50 of A/Vn or vehicle controls (uninfected MDCK cell culture supernatant or allantoic fluid; CC and AF controls, respectively). All challenges occurred in a 50-microliter volume with infectious virus diluted in calcium- and magnesium-free phosphate-buffered saline. Body weights and clinical observations were recorded daily over a 12-day period post-challenge. In order to estimate the 50 % lethal dose (LD50), additional groups of mice were challenged with the A/Vn or Tx/91 viruses using six serial tenfold dilutions or with the 1918 HA/NA:Tx/91 virus in four serial tenfold dilutions in phosphate-buffered saline (4-6 mice for each dose/dilution). The 1918 HA/NA/NS:Tx/91 virus was inoculated into three groups of three mice at a 1 × 106 PFU dose to confirm pathogenicity. The LD50 for the 1918 HA/NA:Tx/91 virus was calculated from a probit dose response model.
For pathogenesis studies, groups of 60-70 mice were challenged with 1 × 106 PFU (Tx/91, 1918 HA/NA:Tx/91, or 1918 HA/NA/NS:Tx/91), 1.5 × 103 TCID50 (A/Vn), or vehicle (controls). The challenge doses for H1N1 viruses were selected based on previously published studies (6). The A/Vn virus is more virulent in mice. The dose for A/Vn was selected to facilitate sample collection through a 120-hour period following infection and provide 100 % mortality with a similar time to death as the 1918 HA/NA:Tx/91 virus. On days 1, 3 and 5 post-challenge mice from each group were sacrificed by cervical dislocation, and the lungs were collected for analysis of gene expression patterns using Affymetrix Gene Chips. Additional mice were sacrificed at each time point for collection of tissues and blood for histopathology and immunoassays. Lungs and blood were pooled from three mice, and a minimum of 2-3 pooled samples were analyzed using 50 % tissue culture infectious dose (TCID50) assays, cytometric bead arrays (CBA; BD Biosciences, San Diego, CA), and hematology and immunophenotyping by flow cytometry.
Blood
Blood was collected by cardiac puncture from groups of three mice into EDTA (K2) vacutainers (BD Biosciences). Four hundred microliters of pooled blood was centrifuged at 500 × g for 5 min at 4 °C. The plasma was transferred to cryovials and stored at -70 °C for CBA assays. The cells were resuspended in an equal volume (400 μL) of stain buffer (BD Biosciences) and stored on ice prior to staining for immunophenotyping. An additional aliquot of pooled whole blood was analyzed using an Advia®120 Hematology Analyzer (Bayer Diagnostics Division, Terrytown, NY).
Lung
Lungs from groups of three mice were homogenized in a single 50-cc tube homogenizer with 1 mL of grinding medium (RPMI-1640 medium containing 1.0 % bovine serum albumin [BSA; Sigma Aldrich, St. Louis, MO] and Benzonase [125 U/mL; EMD Biosciences, San Diego, CA]). The homogenate was centrifuged at 700 × g for 5 minutes at 4 °C. The clarified supernatant was transferred to cryovials and stored at -70 °C for CBA assays, and the cell pellet was resuspended in 1 mL of rinse medium (RPMI-1640 medium supplemented with 125 U/mL Benzonase, 10 % fetal calf serum [Invitrogen, Carlsbad, CA], and penicillin/streptomycin [Invitrogen]). Two milliliters of digestion medium (RPMI 1640 medium with collagenase [200 U/mL; Roche], Benzonase [125 U/mL] and penicillin/streptomycin) was added to the ground lung tissue prior to incubation for 30 minutes at 37 °C in a water bath. This lung digest and the cell suspension from the initial homogenization step were then passed through a 40-micron filter and pooled. The filter was rinsed with 3 mL of rinse medium, and the filtered lung digest was centrifuged at 500 × g for 5 minutes at 4 °C. The final cell pellet was resuspended in 400 microliters of staining buffer and stored on ice prior to staining for immunophenotyping.
Bone marrow
Femurs from groups of three mice were dissected free of tissue, and the ends of the bones were removed using scissors. A 23-gauge needle was inserted into the end of the bone, and the marrow cavity was flushed with 1 to 2 mL of rinse medium. The process was repeated for each end of the femurs. Rinsings collected from the femurs of three mice were pooled in a single 50-mL tube and centrifuged at 500 × g for 5 min at 4 °C. The cells were resuspended in 1 mL of stain buffer and held on ice prior to staining for immunophenotyping.
Immunophenotyping by flow cytometry
For immunophenotyping, pooled lung, blood and bone marrow samples were incubated with three panels of anti-mouse-labeled monoclonal antibodies. A panel of antibodies designed to delineate populations of T-lymphocytes and NK cells (panel 1) included CD49b fluorescein isothiocyanate (FITC), caspase-3 phycoerythrin (PE), CD3 peridinin chlorophyll protein-Cy5.5 (PerCP-Cy5.5), CD45 phycoerythrin-Cy7 (PE-CY7), CD8 Allophycocyanin (APC), and CD4 APC-CY7. Panel 2 (B-lymphocytes and myeloid cells) included CD14 FITC, caspase-3 PE, B220 CD45R PerCP-Cy5.5, CD45 PE-CY7, CD11B APC, and Gr-1 APC-CY7. Panel 3 served to differentiate lineage markers and included TER-119 FITC, B220 CD45R PE, CD3 PerCP-Cy5.5, CD45 PE-CY7, CD11b APC, and APC-CY7. All antibodies were purchased from BD Biosciences (San Jose, CA). Red blood cells were lysed and the cells were washed using an ammonium chloride lysis buffer and stain buffer, respectively. The wash step was repeated, and the stained cells were resuspended in BD Cytofix™ Buffer (BD Biosciences).
Analysis was done using a BD LSR II flow cytometer with FACSDiva™ software version 5.1 (BD Biosciences), and the resultant data were imported into a Microsoft Office Access flow cytometry database for processing. The gating strategy employed targeted specific cell populations by utilizing a combination of DNA content, forward scatter (FS) and side scatter (SSC), as well as CD45 and CD3 staining. Percent population changes from the immunophenotyping assay were converted to actual cell numbers by multiplying total number of lymphocytes from hematology or the total number of white blood cells from the Advia hematology profile by the percentage values obtained for markers in panel 1 or panels 2 and 3, respectively.
Cytometric bead array on lung homogenate and plasma
Levels of interleukin-6 (IL-6), interleukin-10 (IL-10), monocyte chemoattractant protein-1 (MCP-1), interferon-γ (IFN-γ), tumor necrosis factor (TNF), and interleukin-12p70 (IL-12p70) in pooled lung homogenate and plasma samples were measured using the BD™ Cytometric Bead Array (CBA) mouse inflammation assay kit (Becton Dickinson, San Jose, CA) according to the manufacturer’s instructions. The BD™ Cytometric Bead Array Mouse Monokine Induced by Gamma Interferon (MIG, CXCL9) kit was used to evaluate MIG production. Assays were read on a LSR II flow cytometer (BD Pharmingen), and the data were analyzed using the BD Cytomteric Bead Array Software (V 1.4). Comparisons between groups for each respective cytokine/chemokine were performed on log-transformed data using Tukey’s pairwise comparisons with p ≤ 0.05 set as the measure of significance.
RNA isolation, cDNA synthesis, and transcript analysis
Whole lungs were incubated in RNAlater at 2-8 °C for approximately 24 hours prior to transfer to -20 °C. Total RNA was isolated from a minimum of six lung samples for both the control and infected groups using TRIzol® Reagent (Invitrogen, Carlsbad, CA) as per the manufacturer’s protocol. Following chloroform extraction, total RNA was precipitated in isopropanol, washed with ethanol, suspended in nuclease-free water, and stored at –70 °C. The concentration, purity and quality of RNA samples were evaluated using a NanoDrop spectrophotometer (NanoDrop, Wilmington, DE) and a FlashGel RNA cassette system (Lonza, Rockland, ME). Total RNA was used to synthesize double-stranded cDNA using a One-Cycle cDNA Synthesis Kit (Affymetrix, Santa Clara, CA). The cDNA served as a template to synthesize biotin-labeled antisense cRNA using an IVT Labeling Kit (Affymetrix). Labeled cRNA was fragmented and hybridized to the Affymetrix GeneChip® Mouse Genome 430 2.0 Array (containing approximately 34,000 genes) as described in the Affymetrix GeneChip® protocol (Affymetrix). Each microarray was scanned, visualized, and analyzed for the level of each individual transcript using GeneChip Operating Software version 1.2 (Affymetrix).
Analysis of array data methods
Quality control analysis of the arrays was performed using the AffyQC workflow in the BRM (Bioinformatics Resource Manager) software. The AffyQC workflow uses the BioConductor affy package to compute the quality control metrics. The plots that were generated and used to assess the quality of the arrays include a histogram and boxplot of the raw data, RNA degradation plot, 3/5 ratios of QC probesets, RLE boxplot for probesets, and the NUSE boxplot for probesets. Any significant outliers were removed prior to normalization and differential expression analysis.
Normalization and differential expression analysis was performed using the GeneSpring (version 11) software. The arrays were first normalized using the robust multi-array average (RMA) algorithm. Then, the array samples were grouped according to the virus treatment and the corresponding control group. All time points were included in each grouping. A one-way analysis of variance (ANOVA) test was performed on each group to determine differential expression, taking into account unequal variance. Each treated time point was compared to the respective time-matched control. A Tukey’s post-hoc test and the Benjamini-Hochberg method for multiple testing correction were also performed with a false-discovery rate of 0.01. The set of significant, differentially expressed genes were filtered based on a p-value and absolute fold change criterion of 0.01 and 2, respectively.
Histopathology
Additional mice from each group were sacrificed at 24, 72, and 120 hours post-challenge, and the lungs, spleens, liver and brain were removed. Tissues were fixed in 10 % neutral buffered formalin and processed to 5-micron-thick sections. Slides were stained with hematoxylin and eosin (H&E) stain for histopathologic examination by a board-certified pathologist.
Cytospin preparations of lung cells
Lungs were collected from mice 96 hours after challenge with 5 × 104 TCID50 of Tx/91, 1.5x103 TCID50 A/Vn, or H5N1 vehicle control and placed in 0.5 mL of RPMI medium with 0.1 % BSA on ice. The tissues and media were passed through a 40-micron filter into a 50-mL conical centrifuge tube, and the filter was rinsed with an additional 0.5 mL of medium. The rinsings were centrifuged at 700 × g for 5 minutes at 4 °C. The cell pellet and tissue pieces were resuspended in 1.5 mL of digestion medium and incubated for 30 minutes at 37 °C in a water bath. The digest was passed through a 40-micron filter, and the filtrate was centrifuged at 300 × g for 10 minutes at 4 °C. The cell pellets were resuspended in 1 mL of RPMI medium with 0.1 % BSA, and a cell count was performed using a Guava EasyCite (Millipore-Guava Technologies, Billerica, MA). The volume corresponding to 3 × 105 cells was transferred to a separate tube, centrifuged at 300 × g for 10 minutes at 4 °C and resuspended in 1 mL of complete RPMI medium. One hundred microliters of the final cell suspension (30,000 cells) was plated onto slides using a Sukura Cyto-tek Cytospin centrifuge at 1000 RPM for 5 minutes. The slides were fixed in methanol and stained with 4’,6-diamidino-2-phenylindole (DAPI) and polyclonal goat anti-arginase I (Santa Cruz Biotechnology, Santa Cruz, CA) followed by secondary donkey anti-goat IgG Alexa 488 (Invitrogen).
Statistical analysis
For TCID50 and each CBA and immunophenotyping parameter an analysis of variance (ANOVA) model that allowed for unequal variance between challenge group and time points was fitted to the log-transformed assay result. The model fitted to the data was:
where \( y_{ijk} \) is the logarithmic transformed parameter value for the kth BALB/c mouse from the ith challenge group on the jth day after infection, \( \mu \) is the overall mean assay measurement, \( \alpha_{i} \) is the group effect for the ith challenge group, \( \beta_{j} \) is the time effect for the jth day after infection, \( \gamma_{ij} \) is the interaction effect for the ith challenge group and jth day after infection, and \( \varepsilon_{ijk} \) is the residual variation. The model accounts for effects of challenge group and time, as well as the interaction effect between challenge group and time. A graphical assessment of the variability revealed that the assumption of constant variance across groups and time was not reasonable. Thus, a variance structure was chosen that allows for unequal variances between challenge groups and time.
Multiple comparisons were performed to compare each challenge group on each day after infection.
Statistical significance was determined using a Bonferroni-Holm adjustment to correct for the multiple comparisons and control the false-positive rate for the entire collection of comparisons. A corrected p-value less than 0.05 provided statistical evidence that, for a fixed time point, there was a significant difference between the pair of challenge groups.
Results
Viruses containing RNA segments from the 1918 pandemic strain are pathogenic in mice
Viral reassortants carrying the HA, NA and NS genes of 1918 influenza virus were constructed in the H1N1 A/Texas/36/91 (Tx/91) background. The same viral stocks employed by Baskin et al. [12] were used in this study. The viruses expressed the 1918 influenza HA and NA segments or the HA, NA, and NS segments and were designated 1918 HA/NA:Tx/91 and 1918 HA/NA/NS:Tx/91, respectively. The identity of all genes in the reassortants was confirmed by reverse transcription (RT)-PCR and sequencing of the HA, NA, and NS genes. Similar to results obtained by Tumpey et al. [9], viruses containing RNA segments from the 1918 pandemic strain replicated to high titers in Madin-Darby canine kidney (MDCK) cells, similar to those of the parent Tx/91 virus strain (1-2 × 107 PFU/mL for both the 1918 HA/NA:Tx/91 and 1918 HA/NA/NS:Tx/91 viruses).
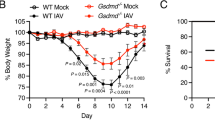
The virulence of the reassortants was determined by challenging BALB/c mice with the viruses by the intranasal route. Female BALB/c mice given an intranasal challenge of 1 × 106 PFU of the 1918 HA/NA:Tx/91 virus lost 23 % body weight by day 5 post-challenge, with all mice succumbing prior to day 11 (Fig. 1a). The 1918 HA/NA/NS:Tx/91 virus which carries the 1918 NS gene in addition to HA and NA was less pathogenic than the 1918 HA/NA:Tx/91 virus. This is consistent with previous results demonstrating that the 1918 NS gene attenuated a mouse-adapted virus in mice, most likely reflecting human adaptation of the 1918 NS1 [7]. Mice challenged with 1 × 106 PFU lost 13 % of their body weight by day 5, with 50 % mortality. Animals challenged with the parent Tx/91 virus displayed no overt disease and gained weight following challenge, and all animals survived. By comparison, the H5N1 A/Vietnam/1203/04 (A/Vn) virus was found to be significantly more pathogenic, consistent with previous literature [13]. An intranasal dose of 1.5 × 103 TCID50 resulted in substantial weight loss by day 5 post-challenge (33 %) and 100 % mortality by day 7. The 50 % lethal dose (LD50) titers in BALB/c mice were 104.14 for 1918 HA/NA:Tx/91 virus, ≥ 106 for 1918 HA/NA/NS:Tx/91, > 106 for Tx/91, and ≤ 1 TCID50 for A/Vn. For comparison, groups of animals challenged with vehicle controls (uninfected MDCK cell culture supernatant or allantoic fluid; H1N1 and H5N1 controls, respectively) survived and gained weight over the course of the study.
Survival and decrease in body weight. BALB/c mice were challenged on day 0 with 1 × 106 PFU of 1918 HA/NA:Tx/91, 1918 HA/NA/NS:Tx/91 or Tx/91; 1.5 × 103 TCID50 of A/Vn; or H1N1 or H5N1 vehicle control administered by the intranasal route. A minimum of six animals were included per challenge group. Values represent (a) mean change from baseline body weight (group averages of pre-challenge weights taken on study day 0), and (b) percent survival by group following challenge
BALB/c mice challenged with 1918 HA/NA:Tx/91 and A/Vn had significantly higher viral burdens in their lungs on days 1, 3 and 5 when compared to mice infected with Tx/91 (Fig. 2). While the viral burden in mice challenged with 1918 HA/NA/NS:Tx/91 was significantly greater than in the lungs of Tx/91-infected animals on day 3, it was not significantly different from that in mice challenged with 1918 HA/NA:Tx/91 or A/Vn at that time, and titers dropped to background level by day 5.
Lung virus titers. BALB/c mice were challenged on day 0 with 1 × 106 PFU of 1918 HA/NA:Tx/91, 1918 HA/NA/NS:Tx/91 or Tx/91; 1.5x103 TCID50 PFU of A/Vn; or H1N1 or H5N1vehicle control administered by the intranasal route. Mice were euthanized on days 1, 3 and 5 post-challenge, lungs from groups of three mice were homogenized, and viral titers were determined by TCID50 assays. Three pooled samples were assayed per challenge group. Values represent mean TCID50 titers per mL (per lung) and the associated confidence intervals. The limit of detection for the assay was 131 TCID50/mL
Infection with A/Vn and 1918 HA/NA:Tx/91 viruses is associated with increased pathology and suppurative inflammation (pulmonary and extrapulmonary)
The histopathology observed in the lungs of infected mice suggested that the H5N1 and 1918 HA/NA:Tx/91 viruses were most similar to one another and appeared to be more pathogenic than the Tx/91 and 1918 HA/NA/NS:Tx/91 viruses (representative images are provided in Fig. 3a–d) (11). Similar to results of previously published studies, the most consistently observed lesions in the lungs of mice infected with highly pathogenic viruses included necrosis and suppurative inflammation (characterized by a neutrophilic response) of the bronchial, bronchiolar, and alveolar epithelium (11). Suppurative inflammation was most pronounced in the lungs of animals infected with1918 HA/NA:Tx/91 and A/Vn. While the characteristics of lesions were not different between infected groups, the relative severity of bronchial and alveolar epithelial necrosis and inflammation in the lungs of mice challenged with A/Vn and 1918 HA/NA:Tx/91 was greater and suggested a longer duration of insult compared to the 1918 HA/NA/NS:Tx/91 or Tx/91 challenge groups. The lung tissues of mice challenged with the vehicle controls were indistinguishable from one another and from the lungs of normal untreated mice.
Pulmonary pathology in mice following intranasal challenge. Alveolar inflammation was generally more severe in mice challenged with 1918 HA/NA:Tx/91 and A/Vn (a and c, respectively) than in mice challenged with 1918 HA/NA/NS:Tx/91 or Tx/91 (b and d, respectively). Black arrows indicate neutrophils in clusters (especially in a, occasionally in c) and singly (b and d) within alveoli. Mice challenged with 1918 HA/NA:Tx/91 and A/Vn also had more striking necrosis of bronchial and bronchiolar epithelium (green arrows). Bronchioli are indicated by a “B”, and vessels are indicated by a “V”. All sections were stained with hematoxylin and eosin stain. 40X original magnification
The progression of pathology was different over time between challenge groups. Bronchial necrosis was present in all influenza-virus-infected mice on day 1. Alveolar necrosis presented early (by day 1) only in mice challenged with Tx/91, though it was of low severity. Animals infected with Tx/91 did not show further progression of lesions. The 1918 HA/NA/NS:Tx/91 virus was slightly more virulent than Tx/91, with alveolar necrosis becoming evident on day 3 and the incidence and severity of alveolar inflammation increasing by day 5. Although the severity of alveolar inflammation increased by day 5, bronchial inflammation had actually decreased for animals challenged with the 1918 HA/NA/NS:Tx/91 virus.
Multifocal suppurative inflammation was occasionally present in the liver of mice infected with 1918 HA/NA:Tx/91 or A/Vn virus (data not shown). Lymphocyte apoptosis and/or necrosis was observed in the bronchial lymph node of mice from the 1918 HA/NA:Tx/91 challenge group on day 3 and in the spleen on day 5 for 1 of 3 mice examined in each case. Lymphocyte apoptosis or necrosis was also observed in the spleen for 1 of 3 mice on day 3, and 2 of 3 on day 5 following infection with the A/Vn virus. Neutrophil accumulation in the spleen, when present, was localized within the red pulp following 1918 HA/NA:Tx/91 infection but was noted within both the red and white pulp for A/Vn-challenged animals. Lymphocyte apoptosis/necrosis and suppurative inflammation were absent in the spleens of animals infected with 1918 HA/NA/NS:Tx/91 and Tx/91.
Cytokine and chemokine production in the lung and peripheral blood following challenge with influenza virus strains
To understand potential influences on immune cell population changes, levels of cytokines and chemokines were measured using cytokine bead arrays (CBA) for lung and plasma samples collected on days 1, 3 and 5 following infection with influenza viruses. Discussion of cytokine and chemokine data is limited to statistically significant observations between virus challenge groups.
IL-6, TNF, and MCP-1 peaked on day 1 in the lung of animals infected with Tx/91 (Fig. 4a). For these animals, the lone chemokine elevated relative to matched vehicle controls after day 1 was MIG. MIG, IL-6 and IFN-γ levels were elevated in animals infected with 1918 HA/NA/NS:Tx/91 relative to seasonal Tx/91 virus on day 5. Similarly, the more-lethal 1918 HA/NA:Tx/91 virus induced higher levels of MIG, IL-6, IFN-γ and MCP-1 relative to the seasonal Tx/91 virus on day 5. A/Vn infection was also associated with significantly greater IL-6, IFN-γ, MCP-1 and MIG production in the lung on days 3 and 5.
Lung and plasma cytokine and chemokine response. BALB/c mice were challenged on day 0 with 1 × 106 PFU of 1918 HA/NA:Tx/91, 1918 HA/NA/NS:Tx/91 or Tx/91; 1.5 × 103 TCID50 of A/Vn; or H1N1 or H5N1 vehicle control administered by the intranasal route. Mice were euthanized on days 1, 3 and 5 post-challenge, and the lungs and blood were pooled from groups of three mice. Cytokine and chemokine levels were determined using cytometric bead arrays. Values represent concentrations (pg/mL) in (a) lung and (b) plasma as determined for 2-3 pooled samples for each challenge group
Systemically, seasonal Tx/91, 1918 HA/NA:Tx/91, and HA/NA/NS:Tx/91 viruses failed to induce the same degree of cytokine and chemokine production in the peripheral blood plasma (Fig. 4b). A/Vn infection was associated with much more pronounced changes in cytokine and chemokine levels in the blood. MCP-1, MIG, IFN-γ and IL-6 were all significantly greater in plasma from A/Vn-infected animals on days 3 and 5 as compared with all other challenge groups.
Infection with A/Vn and 1918 HA/NA:Tx/91 is associated with distinct temporal changes in lymphocyte subsets as well as significant accumulations of CD11b+Gr-1+ cells in the blood, lung and bone marrow
Specific changes in immune cell populations occurring in the blood, bone marrow and lung during influenza virus infection were characterized by flow cytometry. Sub-populations of cells were identified using panels of antibodies designed to delineate T-lymphocyte and NK cell populations, B-lymphocytes and myeloid cells, and a third panel served to differentiate lineage markers.
Mice challenged with 1918 HA/NA:Tx/91 had a transient, albeit significant decline in the number of leukocytes (CD45+ cells) in the peripheral blood on day 3 (Fig. 5a). Significant changes were also observed in peripheral blood T-lymphocytes (CD45+CD3+; T-cells) on day 5 for animals infected with A/Vn, at which time these animals showed tenfold fewer circulating T-cells than any other challenge group.
Characterization of changes in peripheral blood and lung immune cell populations following challenge with influenza viruses. BALB/c mice were challenged on day 0 with 1 × 106 PFU of 1918 HA/NA:Tx/91, 1918 HA/NA/NS:Tx/91 or Tx/91; 1.5 × 103 TCID50 of A/Vn; or H1N1 or H5N1 vehicle control administered by the intranasal route. Mice were euthanized on days 1, 3 and 5 post-challenge, and the lungs and blood were pooled from groups of three mice. Immune-cell populations were identified using a gating strategy that employed a combination of DNA content, forward plus side scatter, and labeling. (a) For peripheral blood, immunophenotyping assay results were converted to actual cell numbers by multiplying values obtained using an Advia clinical hematology analyzer for total white blood cells by the percentages for CD45+ cells (leukocytes) and CD45+CD11b+Gr-1+ cells (granulocytes). Likewise, T-cell subsets were determined by multiplying Advia clinical hematology analyzer values for lymphocytes with the percentage of CD45+CD3+ cells (T cells), CD45+CD3+CD4+CD8- (TH cells), and CD45+CD3+CD4-CD8+ (TC cells) (respectively). (b) For lung, immunophenotyping assay results for the same populations are reported as mean percent values with the associated confidence interval
Consistent with changes in the peripheral blood, A/Vn infection resulted in a significant decline in the proportion of leukocytes in the lung as compared with all other challenge groups on day 5 (41.9 % lower than controls; Fig. 5b). Similarly, the percentage of lung T-cells was also significantly lower on day 5. Among all other groups, the only other observed change in leukocytes and T-cell populations was limited to Tx/91-infected animals, which showed an increase in the proportion of lung CD45+ cells (leukocytes) on day 3 relative to the matched H1N1 vehicle control.
Analysis of T-lymphocyte subpopulations revealed that on day 1, animals infected with 1918 HA/NA:Tx/91 and 1918 HA/NA/NS:Tx/91 had significant decreases in peripheral blood helper T-cells (CD45+CD3+CD4+CD8-; TH; Fig. 5a). T-helper cell populations returned to baseline levels by day 5. In contrast, A/Vn infection resulted in a significantly greater loss of TH cells in the peripheral blood on day 5. Tx/91 infection failed to induce a noteworthy pattern of change in T-helper cell populations.
Increased numbers of cytotoxic T-cells (CD45+CD3+CD4-CD8+; TC) were observed in the peripheral blood on day 1, but not on days 3 and 5, following infection with 1918 HA/NA:Tx/91 and 1918 HA/NA/NS:Tx/91 (Fig. 5a). In contrast, peripheral blood TC numbers in animals challenged with A/Vn were similar to control values on day 1 but then decreased significantly on day 5 post-challenge relative to all other challenge groups. Peripheral blood TC numbers remained similar to controls for Tx/91-infected animals throughout the course of the study.
Infection with influenza viruses resulted in significant increases in the numbers and proportion of CD45+CD11b+Gr-1+ cells across multiple compartments. Cells with this phenotype are consistent with granulocytes. In the peripheral blood, statistically significant increases in CD45+CD11b+Gr-1+ cells were observed in Tx/91-infected animals as compared with A/Vn-treated and vehicle controls on day 1 and with vehicle controls on days 1, 3 and 5 (Fig. 5a). In mice infected with 1918 HA/NA:Tx/91 and 1918 HA/NA/NS:Tx/91, the number of CD45+CD11b+Gr-1+ cells was significantly higher than in A/Vn-infected animals on day 1, Tx/91-infected animals on days 3 and 5, and vehicle controls on days 1, 3 and 5. Increased numbers of CD45+CD11b+Gr-1+ cells in A/Vn-infected animals were observed on day 5 compared with vehicle controls.
Similarly, significant increases in the proportion of CD45+CD11b+Gr-1+ cells were observed in the lungs of animals infected with influenza virus (Fig. 5b). While the proportion of these cells was significantly elevated in the lungs of Tx/91-infected animals on day 3, their numbers declined between days 3 and 5 to a level similar to vehicle controls. This was also observed to a lesser extent for animals infected with 1918 HA/NA/NS:Tx/91 with a maximal increase on day 3, and these animals retained a significantly higher level compared to vehicle controls on day 5. In contrast, the proportion of CD45+CD11b+Gr-1+ cells remained significantly elevated for the 1918 HA/NA:Tx/91 challenge group on days 1, 3 and 5. Significant increases of CD45+CD11b+Gr-1+ cells in the lungs of A/Vn-infected animals were observed on day 3 relative to vehicle controls and on day 5 relative to all other challenge groups.
Changes in the bone marrow included significant increases in the proportion of CD45+CD11b+Gr-1+ cells for 1918 HA/NA:Tx/91–infected animals on days 1 and 5, 1918 HA/NA/NS:Tx/91–infected animals on days 3 and 5, and A/Vn-infected animals on day 5. On day 5, 81 % of all CD45+ cells in the bone marrow of A/Vn-infected mice were CD11b+ and Gr-1+, which is more than 20 % greater than what was observed for Tx/91 and H5N1 vehicle controls at this time (Fig. 6). Seasonal influenza virus (Tx/91) infection did not induce statistically significant changes in CD45+CD11b+Gr-1+ cells in the bone marrow.
Characterization of leukocyte and granulocyte populations in the bone marrow following challenge with influenza viruses. BALB/c mice were challenged on day 0 with 1 × 106 PFU of 1918 HA/NA:Tx/91, 1918 HA/NA/NS:Tx/91 or Tx/91; 1.5 × 103 TCID50 of A/Vn; or H1N1 or H5N1vehicle control administered by the intranasal route. Mice were euthanized on days 1, 3 and 5 post-challenge, and the bone marrow was pooled from groups of three mice. Immune-cell populations were identified using a gating strategy that employed a combination of DNA content, forward plus side scatter, and labeling. Immunophenotyping assay results are reported as mean percent values with the associated confidence interval for (a) CD45+ cells (leukocytes) and (b) CD45+CD11b+Gr-1+ cells (granulocytes)
Global gene expression profile of lung tissue from mice infected with influenza virus
Total lung tissue isolated from infected mice was used for microarray analysis. Expression data were analyzed using Ingenuity Pathway Analysis (IPA; Redwood City, CA). Significantly expressed probe sets relative to matched vehicle control groups as determined by an ANOVA (unequal variance; P < 0.01, Tukey HSD post hoc test and Benjamin Hochberg FDR correction) are summarized in Tables 1 and 2. A comparison of differentially expressed IPA-defined biological functions revealed that on day 1 post-challenge, mice infected with 1918 HA/NA:Tx/91 virus expressed significantly more genes associated with infection and response to infection (respiratory disease, inflammatory response, infection mechanisms, immune cell trafficking, antigen presentation, infectious disease and cell-mediated immunity) than mice infected with the other influenza viruses (Whitney Rank Sum Test p ≤ 0.05). There was no statistical difference in the number of genes expressed in the same functional grouping for Tx/91, A/Vn and 1918 HA/NA/NS:Tx/91 on day 1. All mice that were challenged with influenza virus had significant increases in transcripts for chemokines (CXCL9 and CXCL10), interferon-induced products (interferon-induced proteins 27, 35 or 44 and myxovirus resistance 1 or 2; IFI27, IFI35 or IFI44 and MX1 or MX2, respectively), suppressor of cytokine signaling 3 (SOCS3) and signal transducer and activator of transcription 2 (STAT2). This core set of induced genes is consistent with stimulation of an inflammatory response by influenza virus.
Within the IPA-defined biological function for antigen presentation, A/Vn stimulated more than three times as many CC and CXC chemokine genes as the 1918 HA/NA/NS:Tx/91 virus on day 1 post-challenge (Table 1). A/Vn stimulated the upregulation of CCL2, CCL7, CXCL1, CXCL2, CXCL3, CXCL10 and CXCL11. Additional genes within the IPA-defined biological function for antigen presentation included IL-6, immunity-related GTPase family M (IRGM), signal transducer and activator of transcription 1 (STAT1) and thrombospondin 1 (THBS1). All but CXCL3 were a subset of genes stimulated in lungs of mice infected with the 1918 HA/NA:Tx/91 reassortant. In contrast, mice infected with the 1918 HA/NA/NS:Tx/91 reassortant significantly expressed 10 genes at 24 hours within the IPA-defined antigen presentation function: IL-6, IRGM, chemokines (CXCL9 and CXCL10), Fc fragment of IgG high affinity Ia receptor (FCGR1A), integrin alpha M (ITGAM), ras homolog gene family member B (RHOB), TMP metallopeptidase inhibitor 1 (TIMP1), and vascular endothelial growth factor A (VEGFA). Within this pathway, the chemokines CCL2, CCL4, CCL7, CXCL1, CXCL2 and CXCL11 were all differentially expressed in both the A/Vn and 1918 HA/NA:Tx/91 challenge groups. By comparison, lung tissue from mice infected with the parent strain used to construct the 1918 reassortants significantly expressed only a small subset of the chemokine ligand genes in mice infected with 1918 HA/NA:Tx/91 and A/Vn (CXCL9, CXCL10 and CXCL11).
Infection with pathogenic influenza viruses was found to be associated with significant increases on day 5 of the IPA-defined canonical pathways for arginine and proline metabolism, production of nitric oxide and reactive oxygen species in macrophages, and death receptor signaling (Fig. 7a). Selected differentially expressed (upregulated) transcripts included arginase I (ARG1), arginase II (ARG2), hemoxygenase-1 (HMOX1), cytochrome b-245 beta polypeptide (CYBB), death-domain-associated protein (DAXX) and caspases (CAS3, CAS4, CAS7, and CAS12, respectively) Table 2. Peroxiredoxin VI (PRDX6) and glutathione peroxidase (GPX3) were significantly down-regulated. With the exception of PRDX6, significant changes in expression of these selected transcripts were only observed in the lungs of animals infected with highly pathogenic viruses, A/Vn, and, to a somewhat lesser extent, 1918 HA/NA:Tx/91, particularly on days 3 and 5. Further analysis of the data from day 5 post-challenge using DAVID (the Database for Annotation Visualization and Integrated Discovery) revealed a significant reduction in transcripts within the leukocyte-lymphocyte activation pathway for animals infected with A/Vn.
Differential expression of probe sets for animals infected with influenza viruses. Microarray analysis of whole lung tissue collected from animals on days 1, 3 and 5 following challenge with H1N1 or H5N1vehicle control; 1 × 106 PFU of 1918 HA/NA:Tx/91, 1918 HA/NA/NS:Tx/91 or Tx /91; or 1.5 × 103 TCID50 of A/Vn. The plots illustrate the number of significant genes as compared to matched vehicle controls within selected significant differentially expressed IPA-defined (a) biological function pathways and (b) canonical pathways as determined by an ANOVA with unequal variance and p < 0.01, Tukey HSD posthoc test and Benjamini Hochberg FDR correction and ≥ a twofold change
Infection with 1918 HA/NA:Tx/91 and A/Vn were associated with significant differential expression of a number of functional pathways within the IPA-defined biological function categories for inflammatory response and hematological system development and function including those associated with recruitment of neutrophils, granulocytes, and mononuclear cells. Unique transcripts for 1918 HA/NA:Tx/91 and/or A/Vn infection groups included colony stimulating factor 1 (CSF1), chemokines (CCL4, CCL7, CCL20, CXCL2 and CXCL3,) and myeloid differential primary response gene 88 (MYD88). Additionally, IPA-defined biological function categories associated with recruitment of lymphocytes and homing of bone marrow cells, accumulation of myeloid cells and respiratory burst of phagocytes were uniformly significantly and differentially expressed in the lungs of animals infected with 1918 HA/NA:Tx/91 and A/Vn (Fig. 7b).
Arginase I production in the lungs of animals infected with influenza A virus
Histopathology revealed that infection with highly pathogenic influenza virus resulted in suppurative inflammation in the lung and spleen. Microarray data indicated that arginase I expression was upregulated in the lungs of animals infected with 1918 HA/NA:Tx/91 and A/Vn. Arginase I+ granulocytic cells were present in the lungs of animals infected with influenza virus (Fig. 8a). These changes were pronounced for A/Vn-infected animals (Fig. 8b). By comparison, granulocytic cells isolated from the lungs of animals challenged with alloantoic vehicle control showed only a slight increase in arginase I staining above background levels observed in naïve animals (Fig. 8c and d, respectively). Cells isolated from a naïve animal stained with secondary antibody alone were included as a control (Fig. 8e). While autofluorescence of red blood cells was observed, nucleated cells were uniformly negative for arginase I staining.
Influenza virus infection results in infiltration of arginase-I + cells into the lung. Animals were challenged on study day 0 via the intranasal route with a target dose of 5 × 104 TCID50 of Tx /91, 1.5 × 103 TCID50 of A/Vn, or H5N1vehicle control. Four days post-challenge, lungs were homogenized, and cytospin preparations of isolated cells were prepared. Slides were fixed in methanol prior to staining with DAPI (blue) and anti-arginase (green). Results for (a) Tx/91, (b) A/Vn, (c) vehicle control, and a (d) naïve animal are shown along with (e) cells from a naïve animal stained with secondary antibody alone
Discussion
Infection with highly pathogenic influenza virus has been shown to be associated with intense inflammatory disease including excessive cytokine and chemokine production with accumulation of granulocytes in the lung, as well as systemic leukopenia, lymphopenia, and thrombocytopenia. This response has been characterized by a number of investigators for avian H5N1 viruses, the 1918 H1N1 pandemic virus, and H1N1 viruses possessing the 1918 HA and NA glycoproteins. In the current study, we used a global transcriptomic approach to identify host factors and mechanisms that contribute to a differential innate immune cell response during influenza virus infection. The data from the current study were interpreted in the context of previously published reports.
Tumpey et al. [9] demonstrated that recombinant influenza virus expressing the HA and NA genes of the 1918 virus rescued in the Tx/91 background was lethal without prior adaptation as normally required by human seasonal influenza A H1N1 viruses. Similarly, in this study, while the Tx/91 parental virus did not result in weight loss and did not cause mortality, animals infected with 1918 HA/NA:Tx/91 experienced 23 % weight loss on day 5 and 100 % mortality at a 106 PFU intranasal challenge dose. In a study by Basler et al. [7], 1918 reassortants carrying the NS gene in an A/WSN/33 (H1N1) background were attenuated in BALB/c mice when compared to mice infected with the parent A/WSN/33 virus. Our study confirms the attenuation of virulence of the 1918 HA/NA/NS:Tx/91 strain. Animals infected with 1918 HA/NA/NS:Tx/91 showed 13 % weight loss on day 5 and 50 % mortality at a 106 PFU IN challenge dose. In contrast, A/Vn was extremely pathogenic, causing 33 % weight loss five days post-challenge and 100 % mortality.
Patterns of pathogenicity were mirrored by patterns of viral replication in the lung (A/Vn and 1918 HA/NA:Tx/91 > 1918 HA/NA/NS:Tx/91 and Tx/91). Furthermore, the reduced virulence of the 1918 HA/NA/NS:Tx/91 strain and the seasonal Tx/91 virus was confirmed by lung histopathology. Although bronchial necrosis was observed in all virus-challenged mice on day 1 post-challenge, the degree of necrosis in the lung was less severe in the 1918 HA/NA/NS:Tx/91 and Tx/91 mice. Furthermore, the bronchial necrosis observed in the lungs of Tx/91-challenged mice did not progress on day 3 or 5, and although present on day 3, alveolar necrosis in mice challenged with 1918 HA/NA/NS:Tx/91 resolved by day 5. These results are consistent with the increases in body weight, survival and reduced viral burden observed at that time.
Lung consolidation has been described as a pathological feature of severe influenza virus infection caused by the 1918 pandemic influenza virus and H5N1 virus in humans and in animals [11]. Tumpey et al. found that mice infected with the 1918 HA/NA:Tx/91 reassortant had large pneumonic areas in the lungs with numerous neutrophils in the peribronchium and alveoli [9]. Similarly, in our study, challenge with either 1918 HA/NA:Tx/91 or A/Vn was associated with severe lung pathology including marked necrosis and suppurative inflammation of the bronchial, bronchiolar and alveolar epithelium. A novel observation in this study was multifocal suppurative inflammation in conjunction with lymphocyte necrosis and/or apoptosis in the spleen of 1918 HA/NA:Tx/91 (similar to what was observed for A/Vn infected animals in this study).
Historically, along with severe lung pathology, laboratory findings associated with highly pathogenic avian influenza have included marked lymphopenia and thrombocytopenia [4]. In mice, infection with H5N1 A/Hong/Kong/483/97 virus resulted in an 80 % drop in lymphocytes in the peripheral blood by day 4 post-infection as well as reductions in the numbers of CD4+ and CD8+ cells in the spleen and lung [14]. In the current study, while the seasonal H1N1 virus Tx/91 failed to induce significant changes in leukocyte or lymphocyte numbers, A/Vn infection resulted in leukopenia (CD45+ cells) on day 5 as well as a significant reduction in the proportion of leukocytes in the lung on day 5. A/Vn-infected animals also had significant lymphopenia (CD3+ cells) as well as a significant reduction in the proportion of CD3+ cells in the lung on day 5. Further analysis revealed that a depletion of both TH and TC cells contributed to the observed lymphopenia. A new finding in this study was a significant, albeit transient, reduction in numbers of TH and TC cells in the blood of mice infected with 1918 HA/NA:Tx/91 and/or 1918 HA/NA/NS:Tx/91.
Perrone et al. showed that infection with highly virulent 1918 and H5N1 influenza viruses resulted in significant increases in the number of macrophages and neutrophils in the lungs when compared with mice challenged with either the seasonal human strain Tx/91 or low pathogenicity A/Thailand/SP/83/2004 influenza viruses [11]. It was noted that the numbers of macrophages and neutrophils increased for 7 days following challenge with pathogenic influenza viruses. In the current study, flow cytometry was used to differentiate neutrophils from macrophages/monocytes, including the use of side scatter and cell surface marker combinations. We observed significant increases in the proportion of macrophages in the lungs of A/Vn-infected animals, as well as a significant increase in the number of macrophages in the peripheral blood of animals infected with either 1918 HA/NA:Tx/91 or A/Vn. However, in this study, while temporal differences occurred between groups, significant increases in the number and proportion of CD11b+Gr-1+ cells occurred in the peripheral blood and lung for all influenza challenge groups, regardless of virus strain or degree of pathogenicity.
The question arises as to what drives the observed leukocyte changes in the peripheral blood and lungs of animals infected with influenza virus. Certainly, the extent and distribution of viral replication could contribute to this process. De Jong et al. found that low peripheral blood T-lymphocyte counts and high chemokine and cytokine levels in H5N1-infected individuals correlated with pharyngeal viral loads [3]. In mice, chemokine expression has also been reported to closely follow the level of influenza virus replication and affect the type of inflammatory response to infection [9, 15]. In the current study, BALB/c mice challenged with 1918 HA/NA:Tx/91 and A/Vn were found to have significantly higher viral burdens in their lungs on days 1 and 5. Infection with these viruses was also associated with significantly higher levels of MIG, IL-6, IFN-γ and MCP-1 in the lung, and in the case of A/Vn, in the peripheral blood as well.
Consistent with these observations, microarray data revealed that functional pathways within a number of IPA-defined biological functions relating to recruitment of inflammatory cells (neutrophils, granulocytes and mononuclear cells) were differentially expressed following infection with pathogenic influenza viruses. A total of 20 chemokines (both C-C and C-X-C) were significantly expressed at levels at least twofold higher than the vehicle controls at least one of the three time points examined. Only four were significantly expressed by all groups (CCL5, CCL8, CXCL9 and CXCL10). By comparison, the two most pathogenic viruses, A/Vn and 1918 HA/NA:Tx/91, induced expression of more than two times the number of chemokine genes that were expressed in either the 1918 HA/NA/NS:Tx/91 or Tx/91 group (18 and 16 of the 20 significant transcripts, as compared with 7 and 5, respectively). Twelve of the 20 significant chemokines were limited to the 1918 HA/NA:Tx/91 and A/Vn infection groups. Of these, six were shared by both viruses (CCL2, CCL4, CCL7, CXCL1, CXCL2 and CXCL16), two were unique to 1918 HA/NA:Tx/91 (CCL19 and CCL20), and four were unique to A/Vn (CCL3, CXCL3, CXCL14 and CXCL17). It is interesting that four of the five shared chemokines (CCL2, CCL4, CCL7, CXCL1 and CXCL2) are all associated with chemoattraction, induction or activation of monocytes or neutrophils. Also, for the four chemokines unique to A/Vn, CCL3 (macrophage inflammatory protein-1α), CXCL3, CXCL14 and CXCL17, all are involved in migration, recruitment and/or activation of monocytes or polymorphonuclear leukocytes/granulocytes. These data confirm that along with an increased viral burden in the lung, infection with 1918 HA/NA:Tx/91 and A/Vn are associated with increased expression of chemokines, and the specific chemokines that are differentially expressed may be associated with monocyte and neutrophil accumulation and/or activation.
Infection with highly pathogenic avian influenza virus has been associated with dissemination of the virus to multiple extrapulmonary tissues [20]. Clearly, systemic increases in cytokines and chemokines, as well as disseminated viral replication, could contribute to leukocyte changes in extrapulmonary tissues. Hematopoietic stem cells in the bone marrow give rise to myeloid precursor cells [16]. Under steady-state conditions, these cells generate immature myeloid cells (IMCs) that differentiate into dendritic cells, macrophages and neutrophils. However, pathogen-derived soluble factors or host cytokines can lead to proliferation and activation of the IMC population [17]. For example, during polymicrobial sepsis, signaling through MYD88 has been associated with sepsis-induced CD11b+Gr-1+ cell expansion in the spleen, lymph nodes, and bone marrow of mice [18]. These cells, which accounted for nearly 90 % of bone marrow cells by day 7 after induction of sepsis, were characterized by production of inflammatory mediators and contributed towards T cell suppression. In this study, we used immunophenotyping to examine bone marrow from animals infected with influenza virus in order to determine whether infection alters bone marrow leukocyte populations. Interestingly, we only observed significant increases in the proportion of CD45+CD11b+Gr-1+ cells in the bone marrow of animals infected with pathogenic viruses (1918 HA/NA:Tx/91, 1918 HA/NA/NS:Tx/91 and A/Vn on days 1, 3 and 5, respectively). It has been reported that 20–30 % of the cells in normal mouse bone marrow have this phenotype [13]. In the current study, on day 5, more than 80 % of CD45+ cells in the bone marrow of A/Vn-infected animals were CD11b+Gr-1+.
Cells with a CD11b+Gr-1+ phenotype are consistent with granulocytes and also with myeloid-derived suppressor cells (MDSCs). MDSCs are a heterogeneous population of cells of myeloid origin that consist of myeloid progenitor cells and immature myeloid cells that normally differentiate into granulocytes, macrophages and dendritic cells [reviewed in refs. 16, 17, 19–21]. De Santo et al. [22], demonstrated that infection of mice with influenza A virus (A/PuertoRico/8/34) resulted in the expansion of CD11b+Gr-1+ MDSCs. They further demonstrated the presence of myeloid cells with suppressive activity in peripheral blood leukocytes of humans infected with seasonal influenza A virus. As a potential contributor to lymphocyte dysfunction and depletion, this possibility is significant given that recovery from influenza infection is dependent upon development of virus-specific T and B cell populations [9, 23, 24].
Microarray results in this study demonstrated significant differential expression of pathways associated with respiratory burst of phagocytes and production of nitric oxide and reactive oxygen species. Kash et al. [25] identified a set of genes involved in the production of, and response to, superoxide radicals in the lungs of mice infected with influenza viruses. Consistent with these observations, we found that CYBB, components of membrane-bound oxidase that are involved in the production of superoxide radicals, were up-regulated in the lungs of animals infected with highly pathogenic viruses. We also observed significant up-regulation of transcripts indicative of oxidative damage, including hemoxygenase-1 (HEMOX1), and down-regulation of antioxidants GPX3 and peroxiredoxin VI (PRDX6). These results are consistent with oxidative damage playing a role in the increased pulmonary pathology observed during infection with highly pathogenic viruses. The chemical reaction of NO and superoxide anion can lead to the formation of peroxynitrite in tissues, which has been linked to induced tumor progression and T-cell unresponsiveness [26].
As discussed in a review by Das et al. [27], arginine is a crucial amino acid that serves to modulate cellular immune responses during infection. In the current study, differential expression was observed for the IPA-defined pathway for arginine and proline metabolism. The two isoforms of arginase (ARG1 and ARG2) both hydrolyze the amino acid arginine to ornithine and urea, but they differ with regard to tissue distribution and subcellular localization [24]. L-arginine metabolism by both ARG1 and ARG2 has been implicated in the pathogenesis of asthma and lung inflammation, regulating processes associated with airway tone, cell hyperplasia, and collagen deposition [28]. ARG2 has also been implicated as a potential contributor towards T-cell dysfunction/immunosuppression during cancer [29]. Production of ARG1 by CD11b+Gr-1+ myeloid cells or granulocytes (in mice and humans, respectively) has been shown to result in depletion of L-arginine, which is essential for T-cell activation [17, 19, 30]. In this study, arginase gene expression was most pronounced in the lungs of animals infected with 1918 HA/NA:Tx/91 (ARG2) and A/Vn (ARG1 and ARG2). Arginase I was present in cells in the lungs of animals infected with influenza viruses and was pronounced in lung tissues from animals infected with A/Vn. These results are noteworthy given that microarray data revealed that A/Vn infection was associated with a significant suppression of the leukocyte-lymphocyte activation pathway. Rodrigez et al. [31] demonstrated that low arginine concentration modulates expression of CD3ζ and the T cell receptor. In a subsequent study conducted in our laboratory, microarray data revealed a significant reduction in CD3ζ transcripts in the lungs of A/Vn-infected animals (data not shown).
Taken together, the data demonstrate that inclusion of HA and NA from the 1918 virus renders a seasonal human influenza virus highly pathogenic in BALB/c mice. Similar to H5N1 A/Vn, infection with this reassortant virus resulted in increased viral replication and increased differential expression of chemokines associated with monocyte and granulocyte accumulation and activation. While all animals infected with influenza virus experienced significant increases in CD11b+Gr-1+ cells in the peripheral blood and lung, significant increases only occurred in the bone marrow of animals infected with pathogenic viruses. Notwithstanding the potential impact of these cells on normal hematopoiesis in the bone marrow, the microarray and confocal microscopy data provided in this study suggest that within the lung, the innate immune-cell response results in production of products that have been demonstrated to contribute to T-cell suppression. Additional studies will be required to further characterize the CD11b+Gr-1+ cell response during infection with highly pathogenic influenza viruses.
References
Taubenberger JK, Morens DM (2006) 1918 Influenza: the mother of all pandemics. Emerg Infect Dis 12:15–22
Morens DM, Fauci AS (2007) The 1918 influenza pandemic: insights for the 21st century. J Infect Dis 195:1018–1028
de Jong MD, Simmons CP, Thanh TT, Hien VM, Smith GJ, Chau TN, Hoang DM, Chau NV, Khanh TH, Dong VC, Qui PT, Cam BV, Ha dQ, Guan Y, Peiris JS, Chinh NT, Hien TT, Farrar J (2006) Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat Med 12:1203–1207
Tran TH, Nguyen TL, Nguyen TD, Luong TS, Pham PM, Nguyen VC, Pham TS, Vo CD, Le TQ, Ngo TT, Dao BK, Le PP, Nguyen TT, Hoang TL, Cao VT, Le TG, Nguyen DT, Le HN, Nguyen KT, Le HS, Le VT, Christiane D, Tran TT, Menno DJ, Schultsz C, Cheng P, Lim W, Horby P, Farrar J (2004) Avian influenza A (H5N1) in 10 patients in Vietnam. N Engl J Med 350:1179–1188
Fodor E, Devenish L, Engelhardt OG, Palese P, Brownlee GG, García-Sastre A (1999) Rescue of influenza A virus from recombinant DNA. J Virol 73:9679–9682
Tumpey TM, Basler CF, Aguilar PV, Zeng H, Solorzano A, Swayne DE, Cox NJ, Katz JM, Taubenberger JK, Palese P, García-Sastre A (2005) Characterization of the reconstructed 1918 Spanish influenza pandemic virus. Science 310:77–80
Basler CF, Reid AH, Dybing JK, Janczewski TA, Fanning TG, Zheng H, Salvatore M, Perdue ML, Swayne DE, García-Sastre A, Palese P, Taubenberger JK (2001) Sequence of the 1918 pandemic influenza virus nonstructural gene (NS) segment and characterization of recombinant viruses bearing the 1918 NS genes. Proc Natl Acad Sci USA 98:2746–2751
Watanabe T, Kawaoka Y (2011) Pathogenesis of the 1918 pandemic influenza virus. PLoS Pathog 7:e1001218
Tumpey TM, García-Sastre A, Taubenberger JK, Palese P, Swayne DE, Pantin-Jackwood MJ, Schultz-Cherry S, Solorzano A, Van RN, Katz JM, Basler CF (2005) Pathogenicity of influenza viruses with genes from the 1918 pandemic virus: functional roles of alveolar macrophages and neutrophils in limiting virus replication and mortality in mice. J Virol 79:14933–14944
Pappas C, Aguilar PV, Basler CF, Solorzano A, Zeng H, Perrone LA, Palese P, García-Sastre A, Katz JM, Tumpey TM (2008) Single gene reassortants identify a critical role for PB1, HA, and NA in the high virulence of the 1918 pandemic influenza virus. Proc Natl Acad Sci USA 105:3064–3069
Perrone LA, Plowden JK, García-Sastre A, Katz JM, Tumpey TM (2008) H5N1 and 1918 pandemic influenza virus infection results in early and excessive infiltration of macrophages and neutrophils in the lungs of mice. PLoS Pathog 4:e1000115
Baskin CR, Bielefeldt-Ohmann H, Tumpey TM, Sabourin PJ, Long JP, García-Sastre A, Tolnay AE, Albrecht R, Pyles JA, Olson PH, Aicher LD, Rosenzweig ER, Murali-Krishna K, Clark EA, Kotur MS, Fornek JL, Proll S, Palermo RE, Sabourin CL, Katze MG (2009) Early and sustained innate immune response defines pathology and death in nonhuman primates infected by highly pathogenic influenza virus. Proc Natl Acad Sci USA 106:3455–3460
Cilloniz C, Pantin-Jackwood MJ, Ni C, Goodman AG, Peng X, Proll SC, Carter VS, Rosenzweig ER, Szretter KJ, Katz JM, Korth MJ, Swayne DE, Tumpey TM, Katze MG (2010) Lethal dissemination of H5N1 influenza virus is associated with dysregulation of inflammation and lipoxin signaling in a mouse model of infection. J Virol 84:7613–7624
Tumpey TM, Lu X, Morken T, Zaki SR, Katz JM (2000) Depletion of lymphocytes and diminished cytokine production in mice infected with a highly virulent influenza A (H5N1) virus isolated from humans. J Virol 74:6105–6116
Wareing MD, Lyon AB, Lu B, Gerard C, Sarawar SR (2004) Chemokine expression during the development and resolution of a pulmonary leukocyte response to influenza A virus infection in mice. J Leukoc Biol 76:886–895
Ribechini E, Greifenberg V, Sandwick S, Lutz MB (2010) Subsets, expansion and activation of myeloid-derived suppressor cells. Med Microbiol Immunol 199:273–281
Gabrilovich DI, Nagaraj S (2009) Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol 9:162–174
Delano MJ, Scumpia PO, Weinstein JS, Coco D, Nagaraj S, Kelly-Scumpia KM, O’Malley KA, Wynn JL, Antonenko S, Al-Quran SZ, Swan R, Chung CS, Atkinson MA, Ramphal R, Gabrilovich DI, Reeves WH, Ayala A, Phillips J, Laface D, Heyworth PG, Clare-Salzler M, Moldawer LL (2007) MyD88-dependent expansion of an immature Gr-1(+)CD11b(+) population induces T cell suppression and Th2 polarization in sepsis. J Exp Med 204:1463–1474
Ostrand-Rosenberg S, Sinha P (2009) Myeloid-derived suppressor cells: linking inflammation and cancer. J Immunol 182:4499–4506
Apolloni E, Bronte V, Mazzoni A, Serafini P, Cabrelle A, Segal DM, Young HA, Zanovello P (2000) Immortalized myeloid suppressor cells trigger apoptosis in antigen-activated T lymphocytes. J Immunol 165:6723–6730
Bronte V, Wang M, Overwijk WW, Surman DR, Pericle F, Rosenberg SA, Restifo NP (1998) Apoptotic death of CD8 + T lymphocytes after immunization: induction of a suppressive population of Mac-1 +/Gr-1 + cells. J Immunol 161:5313–5320
De Santo C, Salio M, Masri SH, Lee LY, Dong T, Speak AO, Porubsky S, Booth S, Veerapen N, Besra GS, Grone HJ, Platt FM, Zambon M, Cerundolo V (2008) Invariant NKT cells reduce the immunosuppressive activity of influenza A virus-induced myeloid-derived suppressor cells in mice and humans. J Clin Invest 118:4036–4048
Doherty PC, Allan W, Eichelberger M, Carding SR (1992) Roles of alpha beta and gamma delta T cell subsets in viral immunity. Annu Rev Immunol 10:123–151
Gerhard W, Mozdzanowska K, Furchner M, Washko G, Maiese K (1997) Role of the B-cell response in recovery of mice from primary influenza virus infection. Immunol Rev 159:95–103
Kash JC, Basler CF, García-Sastre A, Carter V, Billharz R, Swayne DE, Przygodzki RM, Taubenberger JK, Katze MG, Tumpey TM (2004) Global host immune response: pathogenesis and transcriptional profiling of type A influenza viruses expressing the hemagglutinin and neuraminidase genes from the 1918 pandemic virus. J Virol 78:9499–9511
Bronte V, Kasic T, Gri G, Gallana K, Borsellino G, Marigo I, Battistini L, Iafrate M, Prayer-Galetti T, Pagano F, Viola A (2005) Boosting antitumor responses of T lymphocytes infiltrating human prostate cancers. J Exp Med 201:1257–1268
Das P, Lahiri A, Lahiri A, Chakravortty D (2010) Modulation of the arginase pathway in the context of microbial pathogenesis: a metabolic enzyme moonlighting as an immune modulator. PLoS Pathog 6:e1000899
King NE, Rothenberg ME, Zimmermann N (2004) Arginine in asthma and lung inflammation. J Nutr 134:2830S–2836S
Tate DJ Jr, Vonderhaar DJ, Caldas YA, Metoyer T, Patterson JR, Aviles DH, Zea AH (2008) Effect of arginase II on L-arginine depletion and cell growth in murine cell lines of renal cell carcinoma. J Hematol Oncol 1:14
Munder M, Schneider H, Luckner C, Giese T, Langhans CD, Fuentes JM, Kropf P, Mueller I, Kolb A, Modolell M, Ho AD (2006) Suppression of T-cell functions by human granulocyte arginase. Blood 108:1627–1634
Rodriguez PC, Zea AH, Culotta KS, Zabaleta J, Ochoa JB, Ochoa AC (2002) Regulation of T cell receptor CD3zeta chain expression by L-arginine. J Biol Chem 277:21123–21129
Acknowledgments
The authors wish to thank Kevin Coty, Lindsay Hendey, Morgan Wendling, Rebecca Migliozzi and Connie Essman-Wood for support with the experiments; Carla Baugess for editorial support; Nick Machesky for microarray support; and Eric Vela for review of the manuscript. These studies were partly funded by NIAID grant P01AI058113 (to AG-S).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Long, J.P., Kotur, M.S., Stark, G.V. et al. Accumulation of CD11b+Gr-1+ cells in the lung, blood and bone marrow of mice infected with highly pathogenic H5N1 and H1N1 influenza viruses. Arch Virol 158, 1305–1322 (2013). https://doi.org/10.1007/s00705-012-1593-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-012-1593-3